This is a new OpenGene project: https://github.com/OpenGene/VisualMSI
VisualMSI
VisualMSI is a tool to detect and visualize microsatellite status from NGS data, by simulating the PCR behavior. VisualMSI extracts the PCR adapters from the reference genome, and tries to map them to the sequencing reads. If the adapters are successfully mapped to a read/pair, its inserted length enbraced by the adapter will be calculated. VisualMSI performs statistics based on the inserted length distribution. This method is very similar as the PCR-based MSI detection method, which is usually considered as the golden standard method for clinical usage.
For each MSI target locus, VisualMSI computes the information entropy of its inserted length distribution. The information entropy value is a indicator for the MSI status, the higher the information entropy is, the higher the probility that this MSI locus is instable.
VisualMSI can run in tumor-normal paired mode or case-only mode, and the tumor-normal mode is suggested if the paired normal sample is available. If the paired normal sample is given, VisualMSI will evaluate the earth mover's distance (EMD) between the distributions of tumor data or normal data. Since the normal data is usually considered as MSI-stable, the EMD value indicates how instable the tumor data is when comparing to the normal data. The higher the EMD value is, the higher probility that this MSI locus is instable.
Take a quick glance of the informative report
- Sample HTML report: http://opengene.org/VisualMSI/msi.html
- Sample JSON report: http://opengene.org/VisualMSI/msi.json
- Sample TEXT report: http://opengene.org/VisualMSI/msi.txt
- Dataset for testing: http://opengene.org/VisualMSI/tumor.sorted.bam and http://opengene.org/VisualMSI/normal.sorted.bam
A quick example
Tumor-normal paired mode:
visualmsi -i tumor.sorted.bam -n normal.sorted.bam -r hg19.fasta -t targets/msi.bed
Case-only mode (no paired normal data given):
visualmsi -i tumor.sorted.bam -r hg19.fasta -t targets/msi.bed
Get visualmsi program
download binary
This binary is only for Linux systems, http://opengene.org/VisualMSI/visualmsi
# this binary was compiled on CentOS, and tested on CentOS/Ubuntu
wget http://opengene.org/VisualMSI/visualmsi
chmod a+x ./visualmsi
or compile from source
# step 1: download and compile htslib from: https://github.com/samtools/htslib
# step 2: get VisualMSI source (you can also use browser to download from master or releases)
git clone https://github.com/OpenGene/VisualMSI.git
# build
cd VisualMSI
make
# Install
sudo make install
Usage
You should provide following arguments to run visualmsi
- the reference genome fasta file, specified by
-ror--ref= - the target setting file, specified by
-tor--target= - the input BAM file, specified by
-ior--in=. If the normal data is available, specify it by-nor--normal= - the plain text result is directly printed to STDOUT, you can pipe it to a file using a
>
Reference genome
The reference genome should be a single whole FASTA file containg all chromosome data. This file shouldn't be compressed. For human data, typicall hg19/GRch37 or hg38/GRch38 assembly is used, which can be downloaded from following sites:
hg19/GRch37: ftp://ftp.ncbi.nlm.nih.gov/sra/reports/Assembly/GRCh37-HG19_Broad_variant/Homo_sapiens_assembly19.fastahg38/GRch38: http://hgdownload.cse.ucsc.edu/goldenPath/hg38/bigZips/hg38.fa.gz Remember to decompress hg38.fa.gz since it is gzipped and is not supported currently.
Target file
The target file is a bed file giving the MSI locuses. VisualMSI will compute the target MSI locus using the center between the start and the end position. To add a MSI target locus at chr:pos, you can add a row with values (chr, pos-100, pos+100, name). You can see the example from targets/msi.bed:
chr4 55588112 55608311 BAT25
chr2 47631460 47651659 BAT26
chr14 23642268 23662467 NR-21
chr11 102183409 102203608 NR-27
chr2 95839262 95859461 NR-24
Please note that this bed file is based on hg19 coordination.
Reports
VisualMSI reports results in HTML/JSON/TEXT formats, you can take a look at following examples:
- Sample HTML report: http://opengene.org/VisualMSI/msi.html
- Sample JSON report: http://opengene.org/VisualMSI/msi.json
- Sample TEXT report: http://opengene.org/VisualMSI/msi.txt
Tumor-normal paired mode
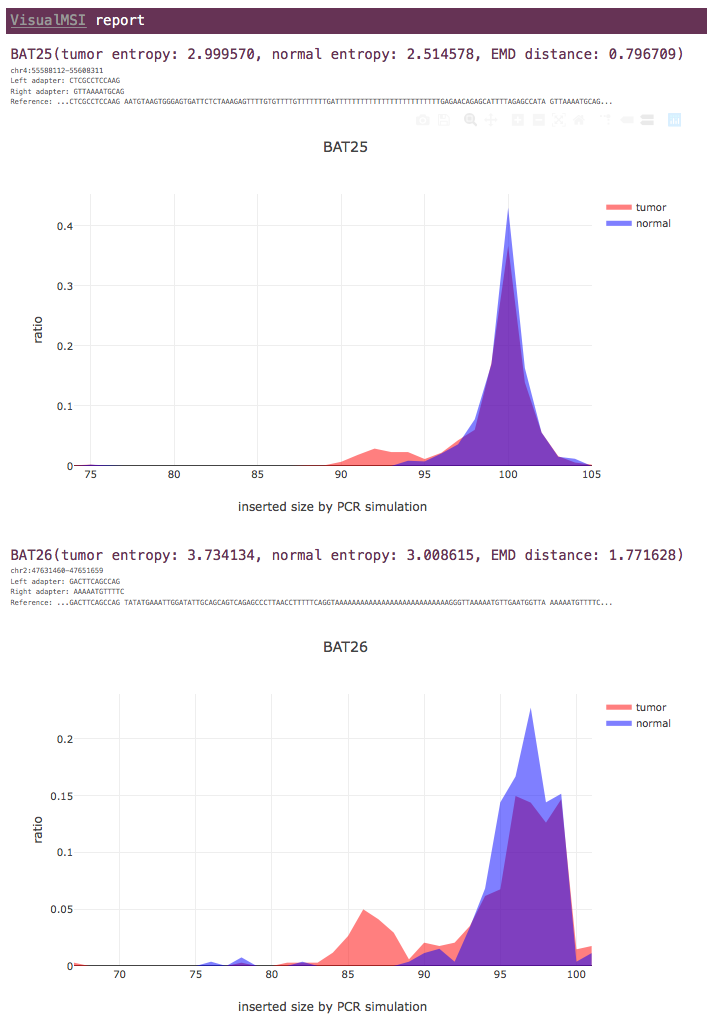
For each MSI locus, the entropy values of tumor and normal data are shown, as well as the earth mover's distance (EMD) value.
Case-only mode
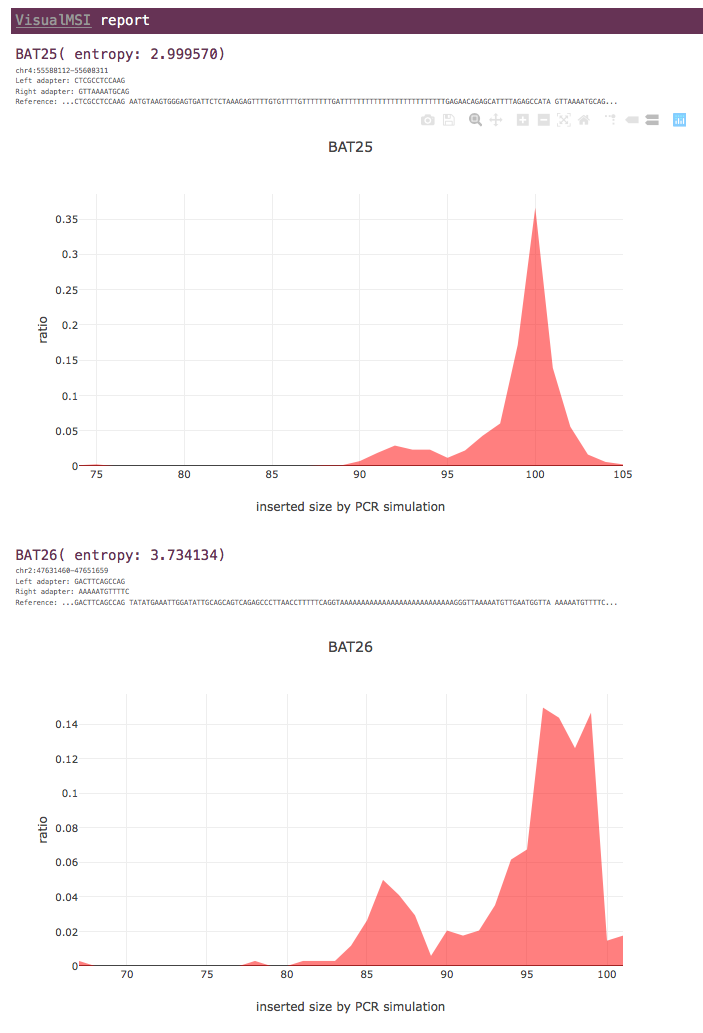
For each MSI locus, only the entropy value of tumor data is shown.
All options
options:
-i, --in input sorted bam/sam file for the case (tumor) sample. STDIN will be read from if it's not specified (string [=-])
-n, --normal input sorted bam/sam file for the paired normal sample (tumor-normal mode). If not specified, VisualMSI will run in case-only mode. (string [=])
-t, --target the bed file (chr, start, end, name) to give the MSI targets (string)
-r, --ref reference fasta file name (should be an uncompressed .fa/.fasta file) (string)
# options for setting thresholds
-a, --adapter_len set the length of the adapter for PCR simulation (5~30). Default 12 means the left and right adapter both have 12 bp. (int [=12])
-l, --target_inserted_len set the distance on reference of the two adapters for PCR simulation (20~200). Default 100 means: <left adapter><100 bp inserted><right adapter> (int [=100])
-d, --depth_req set the minimum depth requirement for each MSI locus (1~1000). Default 10 means 10 supporting reads/pairs are required. (int [=10])
# options for specifying the file names of the reports
-j, --json the json format report file name (string [=msi.json])
-h, --html the html format report file name (string [=msi.html])
# other options
--debug output some debug information to STDERR.
-?, --help print this message


Hi Chen, I tried downloading/saving VisualMSI using the steps given above, but when I test the tool in command line, it says visualmsi: command not found
Could you please help me on this?
TIA
Hey. It is mentioned that the target file is based on hg19 coordination. How can I generate one for hg38 coordination? TIA
You can make your own file if you want to process hg38 bam.