The GenomeGraphs package in Bioconductor allows to draw (human) chromosome ideograms with R. The package can be used to depict genome tracks of coverage, microarray measurements and genes together with the ideograms. See the user guide for an overview of different types of graphics. For the ideogram, the example looks like this (also in the GenomeGraphs paper):
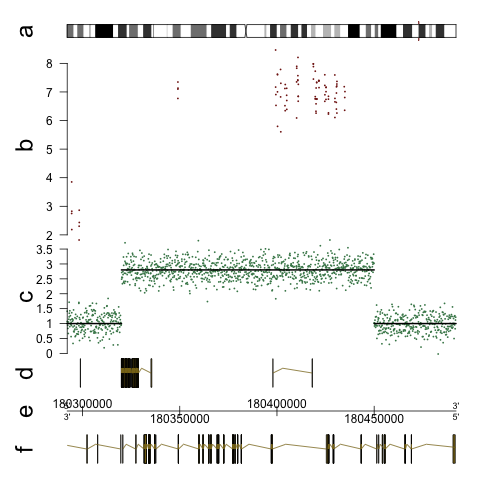
Here is the code that makes something like this (from the user guide):
library(GenomeGraphs)
library(biomaRt)
data("exampleData", package = "GenomeGraphs")
mart <- useMart("ensembl", dataset = "hsapiens_gene_ensembl")
minbase <- 180292097
maxbase <- 180492096
genesplus <- makeGeneRegion(start = minbase,
end = maxbase, strand = "+", chromosome = "3",
biomart = mart)
genesmin <- makeGeneRegion(start = minbase,
end = maxbase, strand = "-", chromosome = "3",
biomart = mart)
seg <- makeSegmentation(segStart, segEnd,
segments, dp = DisplayPars(color = "black",
lwd = 2, lty = "solid"))
cop <- makeGenericArray(intensity = cn,
probeStart = probestart, segmentation = seg,
dp = DisplayPars(size = 3, color = "seagreen",
type = "dot"))
ideog <- makeIdeogram(chromosome = 3)
expres <- makeGenericArray(intensity = intensity,
probeStart = exonProbePos,
dp = DisplayPars(color = "darkred",
type = "point"))
genomeAxis <- makeGenomeAxis(add53 = TRUE,
add35 = TRUE)
gdPlot(list(a = ideog, b = expres, c = cop,
d = genesplus, e = genomeAxis, f = genesmin),
minBase = minbase, maxBase = maxbase,
labelCex = 2)
Edit: It supports multiple ideograms in one plot like this:
ideog <- makeIdeogram(chromosome = 1)
ideog2 <- makeIdeogram(chromosome = 2)
ideog3 <- makeIdeogram(chromosome = 3)
ideog4 <- makeIdeogram(chromosome = 4)
gdPlot(list("1"= ideog, "2" = ideog2, "3" =ideog3, "4"=ideog4 ),
minBase = minbase, maxBase = maxbase)
If you plot data below the chromosomes using a base track, take care of the minbase, maxbase parameters because the chromosomes have different length!




Dear all, I would like to plot ideogram of zebrafish or stickleback. I tried some of your suggestions here. But they are mainly in human. Do you have any other suggestions? Regards, Nima
Nima - please post this as a new question.
there is a post here: (Creating chromosome karyotype plot with R and ggplot2) which might help get you started, it does not currently include the cytobands but you can add them with the file
cytoBand.txt.gzfrom here