Dear Colleagues,
I would like to introduce Venice, a non-parametric approach for finding marker genes in single-cell RNA-seq data, developed by BioTuring team. Using a widely adopted benchmarking approach (Wang et al. 2019), Venice obtains the best accuracy compared with 14 popular methods, while keeping a modest running time.
For preliminary benchmark, please visit here: https://blog.bioturing.com/2019/06/24/venice-a-non-parametric-test-for-finding-marker-genes-in-single-cell-rna-seq-data/
Venice is open-source, and freely available for academic entities. The method is now incorporated in Signac, a single-cell analytics package developed by BioTuring, available at https://github.com/bioturing/signac.
Some benchmark results:
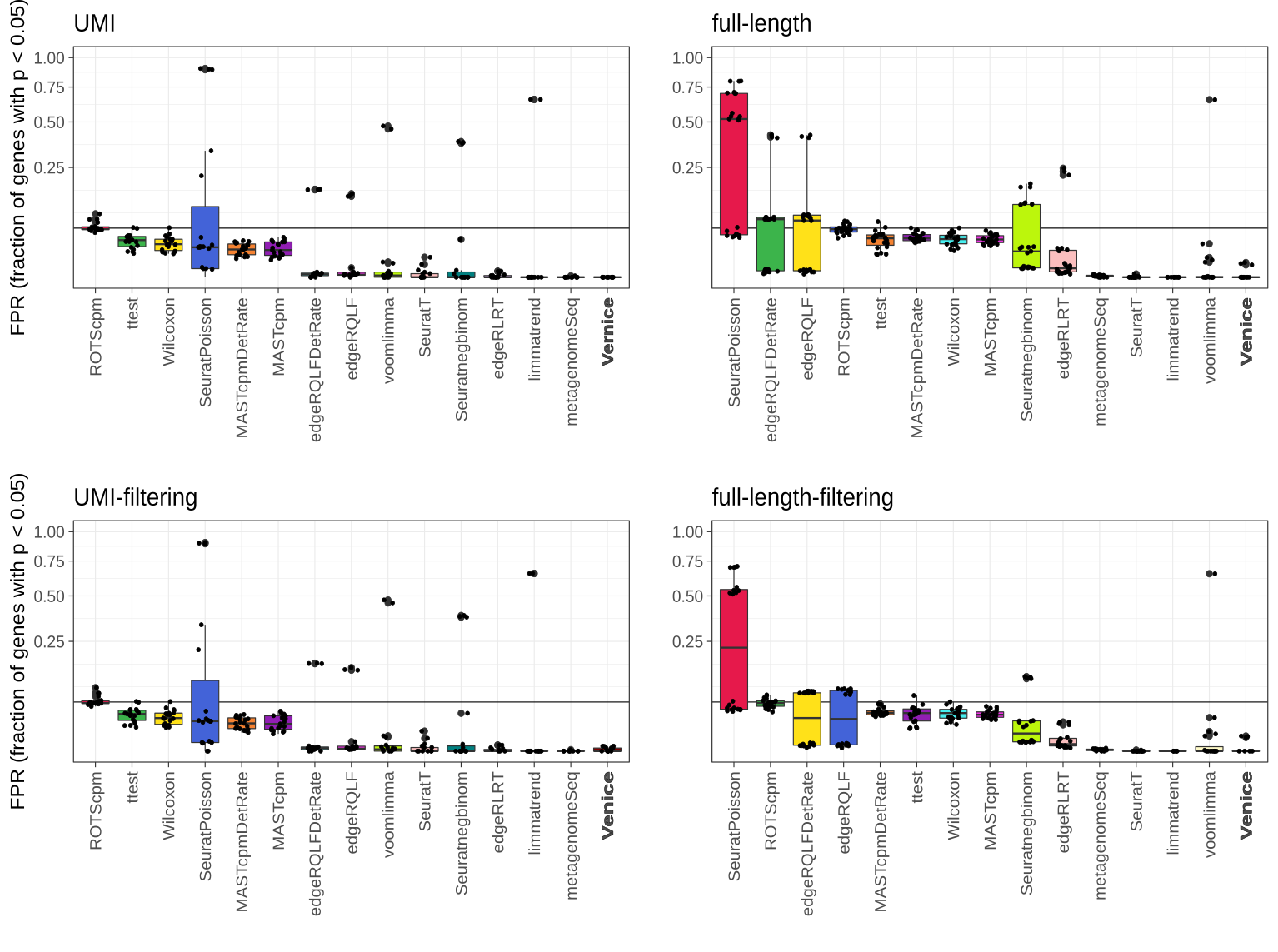
Type I error control across 87 instances from eight real single-cell null data sets. The black line indicates the target FPR = 0.05 and the y-axis is square-root transformed for increased visibility. Centerline, median; hinges, first and third quartiles; whiskers, most extreme values within 1.5 interquartile range (IQR) from the box. The benchmark was adopted from Sonesson et al. (2018).
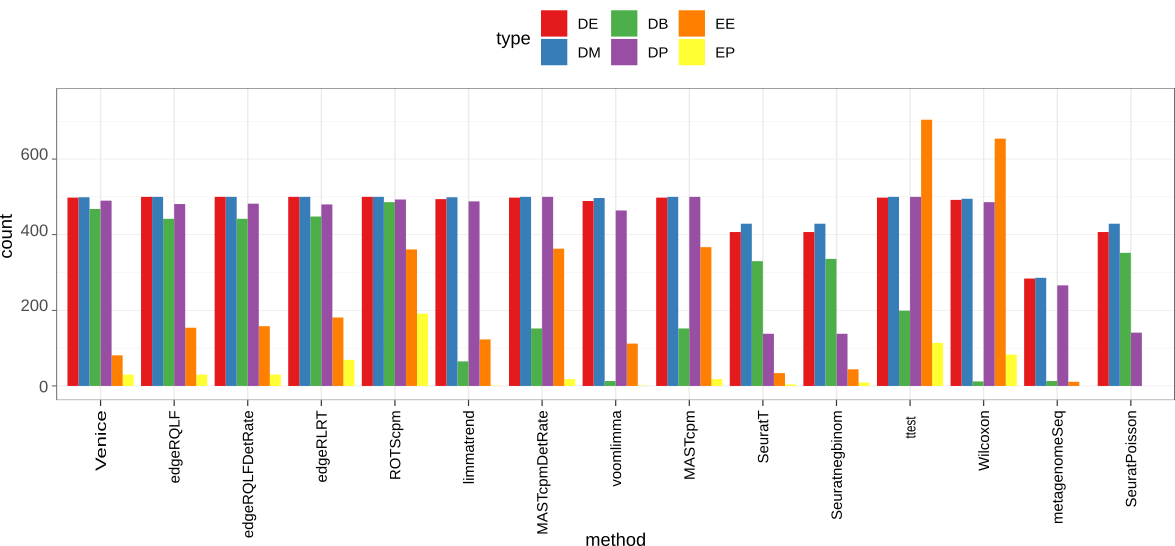
Accuracy on the simulated data. We use scDD package to simulate a dataset contains 2000 differential expressed genes (DEG) that span across four groups: DE, DB, DM, DP. The dataset also has 18000 non-DEG of types EE or EP.
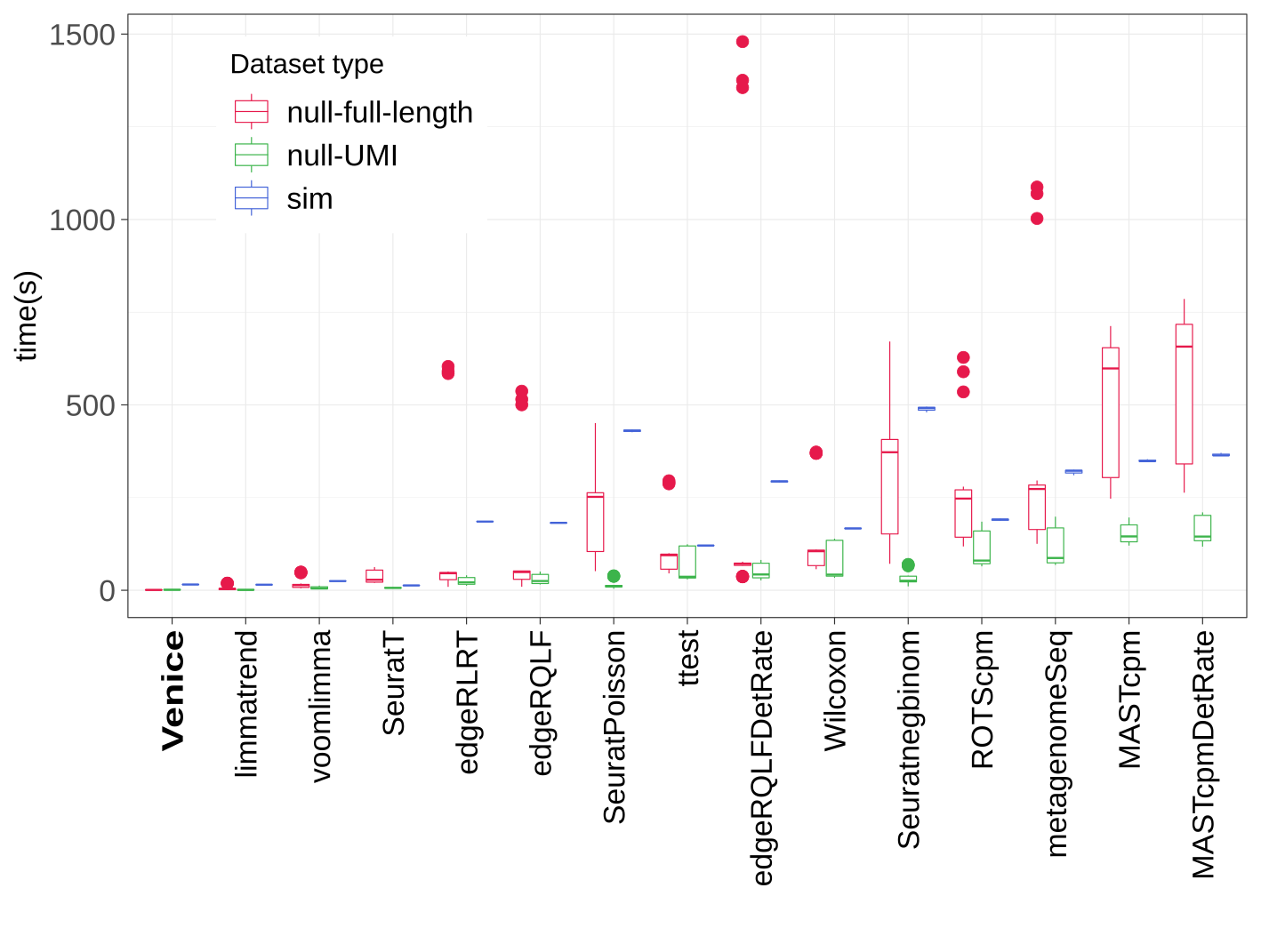
Running time (in seconds) of all methods.

