Entering edit mode
5.5 years ago
zhaoliang0302
▴
50
Hi,
I detected several outliers among my samples by plotting PCA. But I don't know how to remove this samples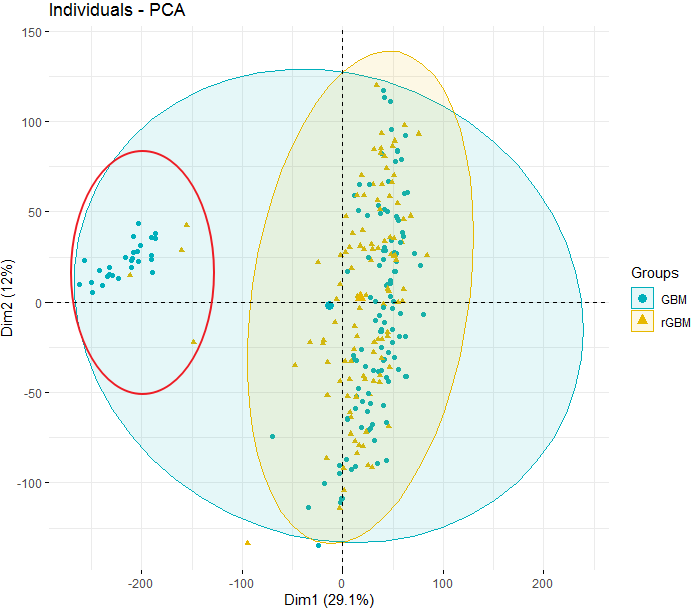 The outlier samples is marked by the red circle.
The outlier samples is marked by the red circle.
Thanks


You should explain how you generated your PCA plot (from which type of data ? ). Put your code. And a minimal reproducible example.
The data is a dataframe of RNAseq FPKM expression file, rows correspond to genes and columns to samples.
My first question with such a plot is, what are these outlier samples? Is there a biological or technical explanation for this?
I downloaded this RNAseq data and just explore it. Considering the large samples, I think remove these 'outlier' samples is not a risk.
Are all samples from the same dataset ? Do you have metadata on this samples (sequencing kit ? type ? cell type ? sequencing plateform, etc...) IMO you see here a clear (non-biological) batch effect
Yes, all tumor samples are from the same dataset. The clinical data doesn't contains batch information. So I want to remove these samples directly.
I guess in the
pcaobject you should havePC1andPC2(information used to plot). Use these to filter out the samples i.e.PC1 < -100Thanks, I save this plot as PDF file (large size) and then zoom in to get the outlier samples. It sounds silly but it really works :-)
Hi, I am also facing the same issue, and by checking your suggested method I am finding the actual sample which have pc1 < -100 are outlier. Please can you share explanation what is the basis of the threshold selection of -100. It would be much helpful. Thank you.
ues fviz_pca_ind(pca_all, geom.ind = "text") to show sample names on the plot.