Hi,
I have read in a few places that one needs ~10-20 million reads per sample to do differential gene expression analysis (see Harvard RNA-seq Tutorial,"Improving mean estimates (i.e., reducing variance) with biological estimates" section as an example).
However, I'm wondering does this refers to the total number of reads or the number of reads which fall within the coding portions of the genome. I have seen samples which have ~80 million reads, but when looking at count data in DESeq2, the sum of counts for the sample in question only has a count sum of ~3 million.
So, I'm curious how to interpret this ~10-20 million reads per sample rule-of-thumb.
Thanks

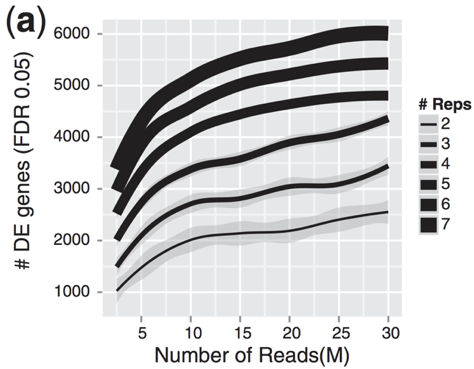

I normally expect between 1/3 and 2/3 of reads sequenced to fall into exonic regions (depending on many thing, but primarily if the sample is total RNA or polyA RNA). There is something weird about a sample where only 3/80 million reads map to exons.
Also, I'd say 10-20 million reads is a bit on the low side.