Entering edit mode
5.1 years ago
Shicheng Guo
★
9.6k
Hi All,
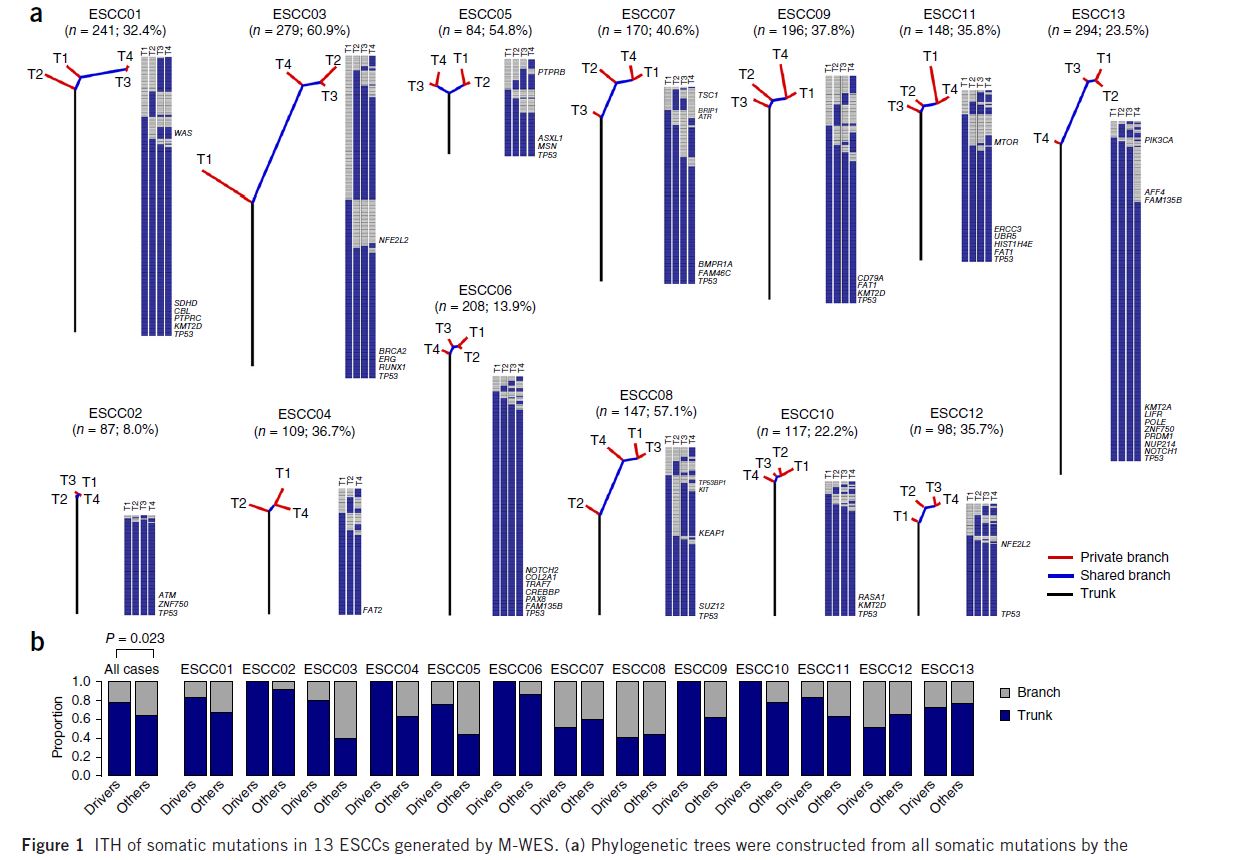
I want to build a tree to reflect the relationship between multiple samples with the following style (Figure 1a, Nature Genetics, PMID: 27749841). Suppose I have a 0,1 matrix, rather than DNA sequences/variants matrix, as the right panel of the phylogenetic tree, is there any method to build a similar figure? R or Python package is preferred. I am not sure whether mega 7 can show the tree like this style.
Thanks.
Phylogenetic trees were constructed from all somatic mutations by the Wagner parsimony method.


Great. I will check these two tools. Thanks for the suggestion.