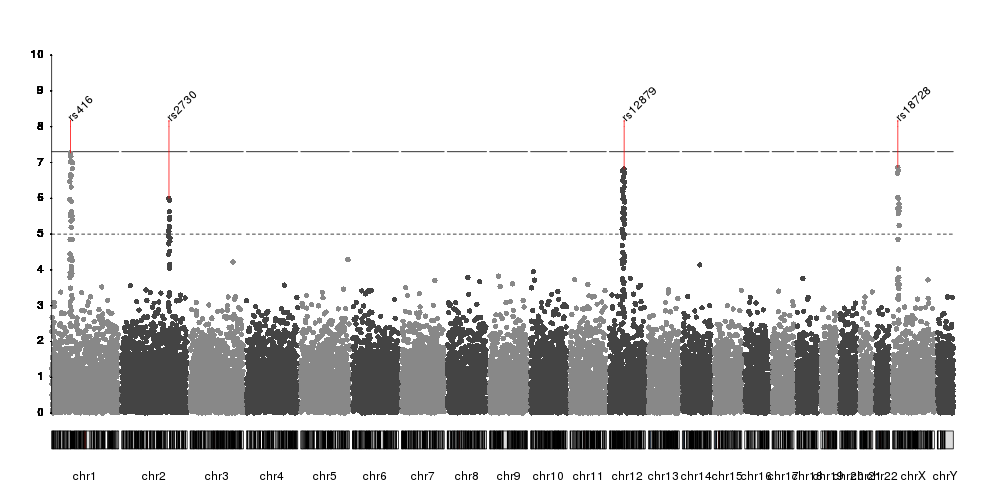
Hello everyone? I wonder if someone knows how to change the chromosome labels at qqman package? I am making a manhattan plot using manhattan function which adds numeric chromosome codes to the x-axis like this:

However, the species under study has 2 subgenomes (A and C) so the chromosome codes should look like that: A1, A2... and C1, C2... And the manhattan x-axis would be like that:
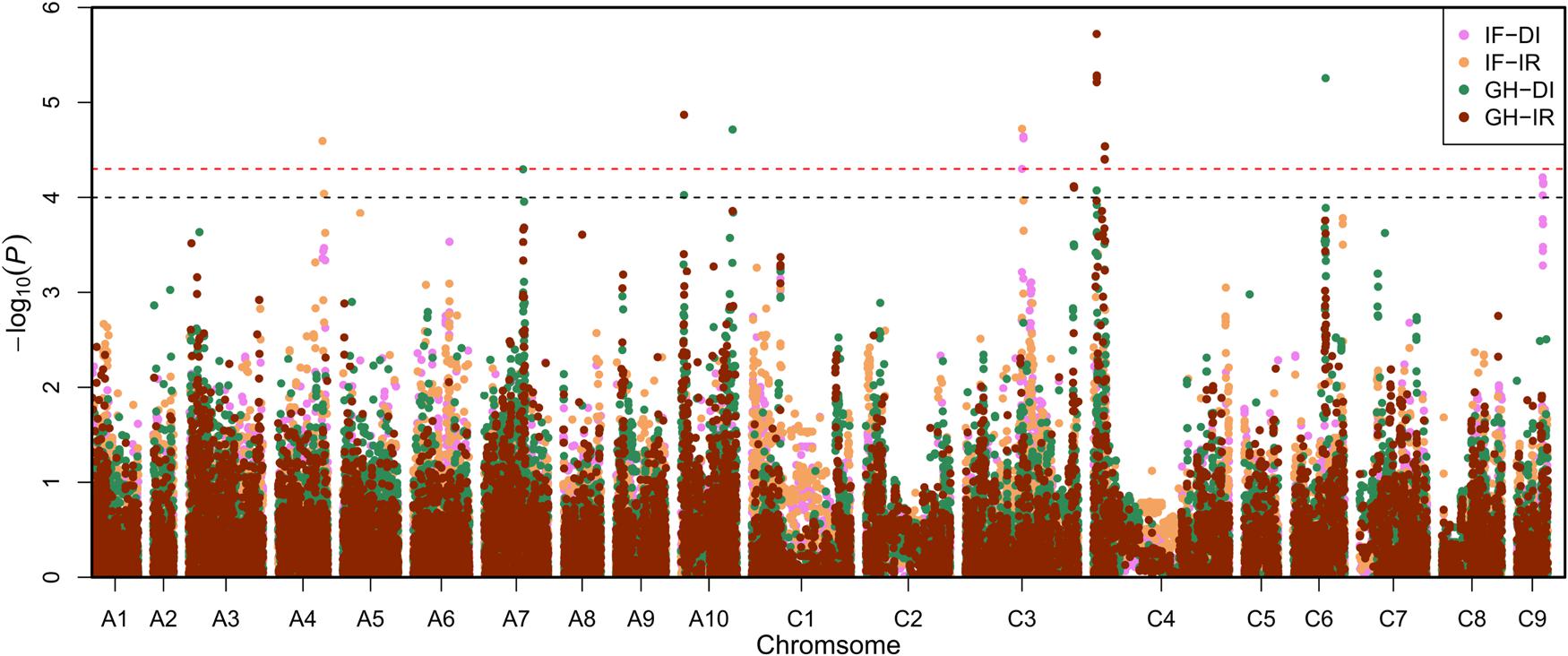
I didn't find the solution for such a problem, so please, help.