Hi,
I have downloaded some public data PRJNA427246 from SRA database. The paper is “Hybrid Sequencing Reveals Insight into Heat Sensing and Signaling of Bread Wheat.”. https://doi.org/10/ggffjg. The sra data information are as follow:
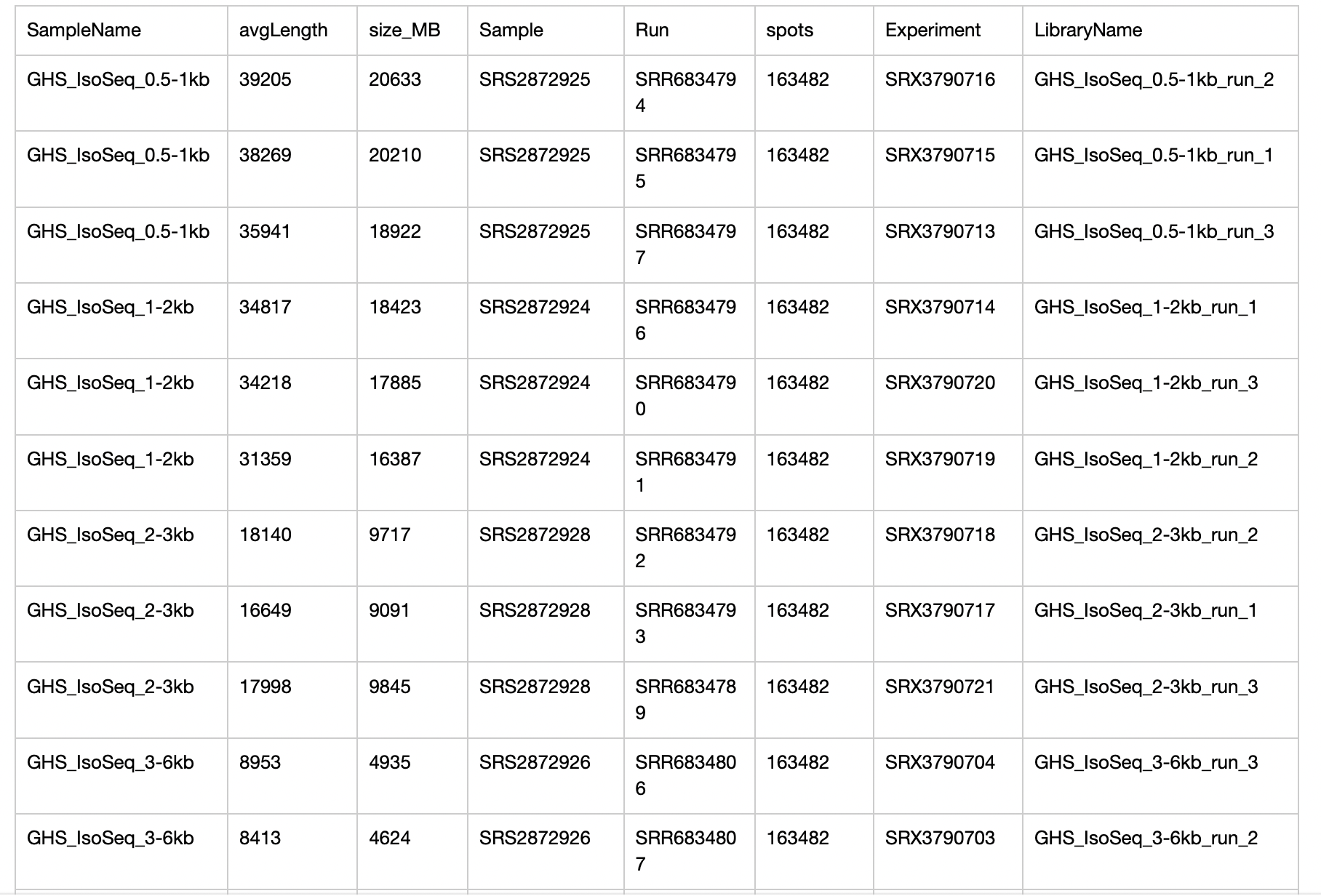
When the download is complete, I convert the sra files into fastq files use fastq-dump. Now I want to analyze these fastq data, however, many analysis pipeline and methods are start from bam file. I would like to ask how to start the analysis from the fastq file downloaded from SRA database.
Best wishes


the pacbio bam file have many orther information, just cnvert fastq file in to sam file?