Hello all,
Please bear with me, I have very little experience with bioinformatics and I'd really appreciate everyone's help with my questions.
I have some confusion regarding gene ontologies and the genes associated. I have some gene ontology terms enriched from a ChIP-seq experiment, and I'd like to be able to classify whether such genes belong to different categories.
While I understand that HOMER will check different libraries of gene grouping, like GO, COSMIC etc, then rank them according to p-values, I am unclear about what it means by 'Common Genes', and why each 'Term' has multiple genes, especially when row 1 and row 2 look to be related processes:
Picture from HOMER website: 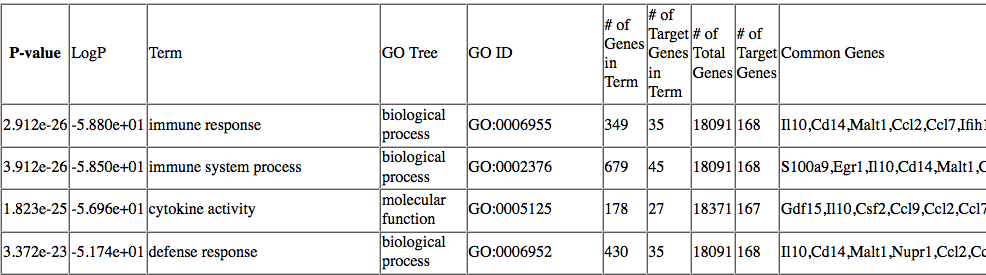
Is it possible that I can use each of these Common Genes and categorize them accordingly (e.g. inflammatory genes, apoptosis signalling genes etc.)? If so, is there a bioinformatics tool that I use to categorize these genes, or some sort of database that I can use for inflammatory genes and match them up for all the Common Genes in each Term? Alternatively, does HOMER already 'classify' these genes for me through this gene enrichment?
I ask this since my significant Terms are from COSMIC (e.g. 'blood_vessel' or 'prostate'), and COSMIC is for somatic mutations in human cancer, which is not necessarily what I am studying.
Thanks so much!


Thanks so much for the very detailed workflow! Really appreciate the time you took to help me out; I will try this out and let you know if I have any more questions.