Hello Biostars, I am working on the genome analysis of Rousettus_aegyptiacus organisms from data obtained ENA database. After the quality checking (Fastqc) of the reads, I have done alignment with hiasat2, index building carried out using the file from NCBI: ftp://ftp.ncbi.nih.gov/genomes/Rousettus_aegyptiacus/CHR_Un/9407_ref_Raegyp2.0_chrUn.fa.gz. Then I tried to check the overall alignment quality score for a few samples, it ranges 92-93%. Now I want to perform stringTie for assembly, In this step, I need the GTF file. I couldn't find it. Please tell me how do I get or generate GTF fie for this organism. Your response highly appreciated! Thank you
You can download the GTF file from NCBI by searching for Rousettus aegyptiacus in the NCBI Assembly portal, select the top hit use the blue Download button as shown in the image below:
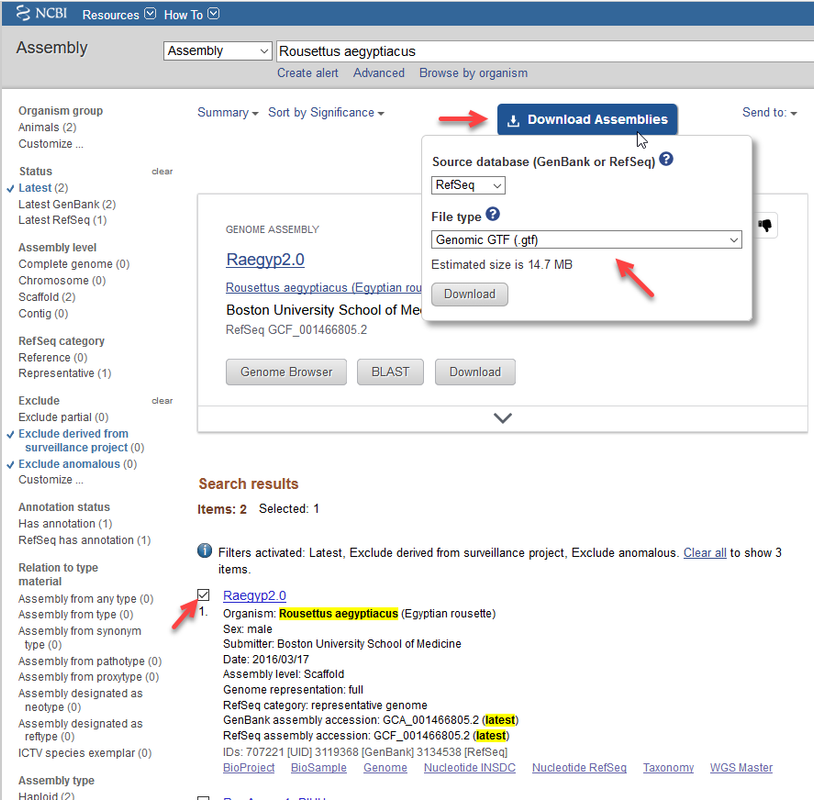
Thank you very much for the response Response from stringTie Please make sure the -G annotation file uses the same naming convention for the genome sequences.
I generated hisat index files using the fasta file from the link (https://www.ncbi.nlm.nih.gov/assembly/?term=Rousettus+aegyptiacus) image file also attached ![enter image description here][1]
pl reply me
It looks like the annotation file and the genomic FASTA files do not have matching genomic seq-ids. If you use the following two files, I expect the seq-ids to be matching:
Genomic FASTA: ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/466/805/GCF_001466805.2_Raegyp2.0/GCF_001466805.2_Raegyp2.0_genomic.fna.gz GTF: ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/466/805/GCF_001466805.2_Raegyp2.0/GCF_001466805.2_Raegyp2.0_genomic.gtf.gz
You can check which genomic seq-ids were used for the hisat2 index using the hisat2-inspect command.
a GFF is available at: ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/001/466/805/GCF_001466805.2_Raegyp2.0/GCF_001466805.2_Raegyp2.0_genomic.gff.gz and then converting .gff file to .gtf
make sure that you use gtf/gff3 and reference from the same source. @ mathavanbioinfo
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


https://ibb.co/1KQbrB7
This is incorrect. Link is about reference (fasta) downloading image. OP is about annotation (GTF/GFF3) file.