Hi, I'm sorry for my ignorance, but I'm still a student. How could I find the target genes of a specific transcription factor? I also need to evaluate if the regulated target genes are the same in two different conditions. Thank you so much for your help!
To build upon Mensur's very informative answer, the ideal scenario would likely be to perform RNA-seq on both wild-type cells and cells with the TF of interest knocked out or otherwise repressed. This could be accomplished via a CRISPR-Cas9 mediated deletion of the TF's TSS/first exon or via RNA interference. The former is more full-proof than the latter, though the latter could work well (and is generally easier/quicker).
You could then compare gene expression between the two conditions (wild-type and KO) to identify the genes whose expression is altered - these would likely be direct or indirect target genes of your TF of interest. This could be coupled with ChIP-seq in the wild-type condition to identify regulatory elements where the TF binds directly (which provides evidence of direct targeting, rather than modulating expression of another transcriptional regulator that targets said gene).
Identifying true gene targets is challenging, and incorporating as much information as possible (like chromatin interactions/architecture via Hi-C/5C) can provide additional confidence in your predictions. Biology also plays a big role - if your TF binds almost exclusively at promoters, then the entire process is a lot less challenging. Conversely, if it binds more heavily at distal regulatory elements, finding true target genes can be very difficult, particularly in gene-rich regions.
Transcription factors (TFs) typically bind regulatory DNA elements upstream of a gene (promoters), and by doing so either prevent its transcription, or recruit other proteins that will increase the transcription. There are no simple rules as to what is or isn't a valid regulatory element, though in prokaryotes they tend to be palindromic. It is important to understand that TFs don't bind with absolute specificity, meaning that they are not looking only for one sequence and ignoring everything else. Instead, TFs have strict requirements only in some parts of their recognition elements, and are less stringent about the others. This is reflected in the sequence logo below that shows a compressed representation for a collection of binding sites recognized by LacI. When you align a large number of LacI target sites, a pattern emerges showing what positions within the element are (near-)perfectly conserved, and also what is the likelihood of the four nucleotides at any position.
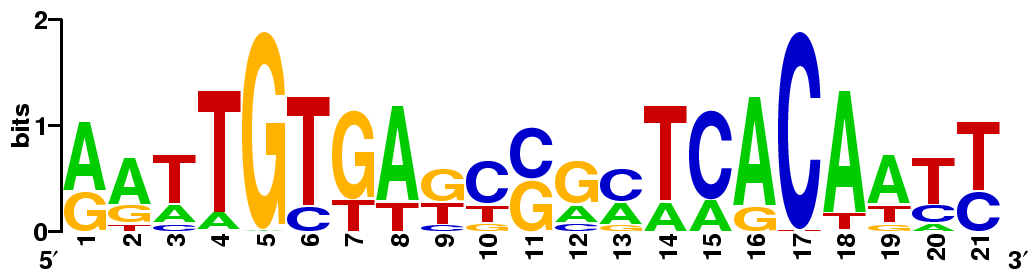
For example, if you pick a tallest letter from each position in the logo above, you would end up with AATTGTGAGCCGCTCACAATT, which is likely to be a very good target for LacI. If you pick the shortest letter from each column, or even letters that are not there at all, say CCGGTACCAAACGGGTGCCGG, that would likely be a terrible target that LacI will ignore. You can read more about sequence logos or even make them here. By the way, the logo above to a trained eye is immediately palindromic and recognized by a dimeric TF, but eukaryotic TFs tend to bind as monomers and do not recognize palindromic elements.
Since there is a continuum of sequences recognized by TFs, one needs to know at least several genes that are recognized by it. From there, computers can identify over-represented DNA elements in promoters by motif sampling, which is how that LacI logo was created. There are databases of various TF recognition elements that Kevin pointed out to you, and their matrices can be used to scan any DNA sequence for matches in sliding window fashion. Motif matrices are probabilistic representation of the information displayed above in the sequence logo, and will give you a score reflecting how well a piece of DNA matches known or predicted binding preferences of a given TF.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Do not worry - many students know more about civility, life, and research than even Full Professors. Popular databases include:
It seems that you have already generated some data, though (?) - can you describe the data that you have produced.
I'm sorry, I'm afraid I didn't explain myself well. I have to design an experiment in order to identify the unknown target genes of a specific transcription factor. I've thought about ChIP-seq, but in that way I would identify the transcription factor binding sites, and I don´t know how to use that info to identify the target genes. Besides, I need to check if the regulated genes are the same in two different conditions. Thank you so much for your attention! You are both really helpful!
No problem. You now have 2 comprehensive answers (see below)