Hello, I am having a difficult creating a visual/ graph to represent my bedgraph. My bedgraph was made from intersecting a bed file (containing about 50 different contig names) with a bam file obtained from mapping small RNA with bowtie. I want to create a bedgraph plot with this information in R file.bg) to represent all the different contig. Is there a way I can do this or any better suggestion on what I can do?
Here is a few lines from my bg.
V2_5 124587 124588 1908
V2_5 124588 124589 1551
V2_5 124589 124590 1733
V2_5 124590 124591 1898
V2_5 124591 124592 2107
V2_18 834476 834478 424
V2_18 834478 834479 325
V2_18 834479 834480 205
V2_18 834480 834482 136
V2_18 834520 834524 55
V2_18 834524 834525 86
V2_18 834525 834526 87
V2_18 834526 834528 88
V2_25 2534300 2534302 240
V2_25 2534302 2534308 260
V2_25 2534308 2534322 263
V2_182 605998 605999 614
V2_182 605999 606000 590
V2_182 606000 606001 422
V2_182 606001 606002 335


If it's possible please provide a few lines of your bedgraph.
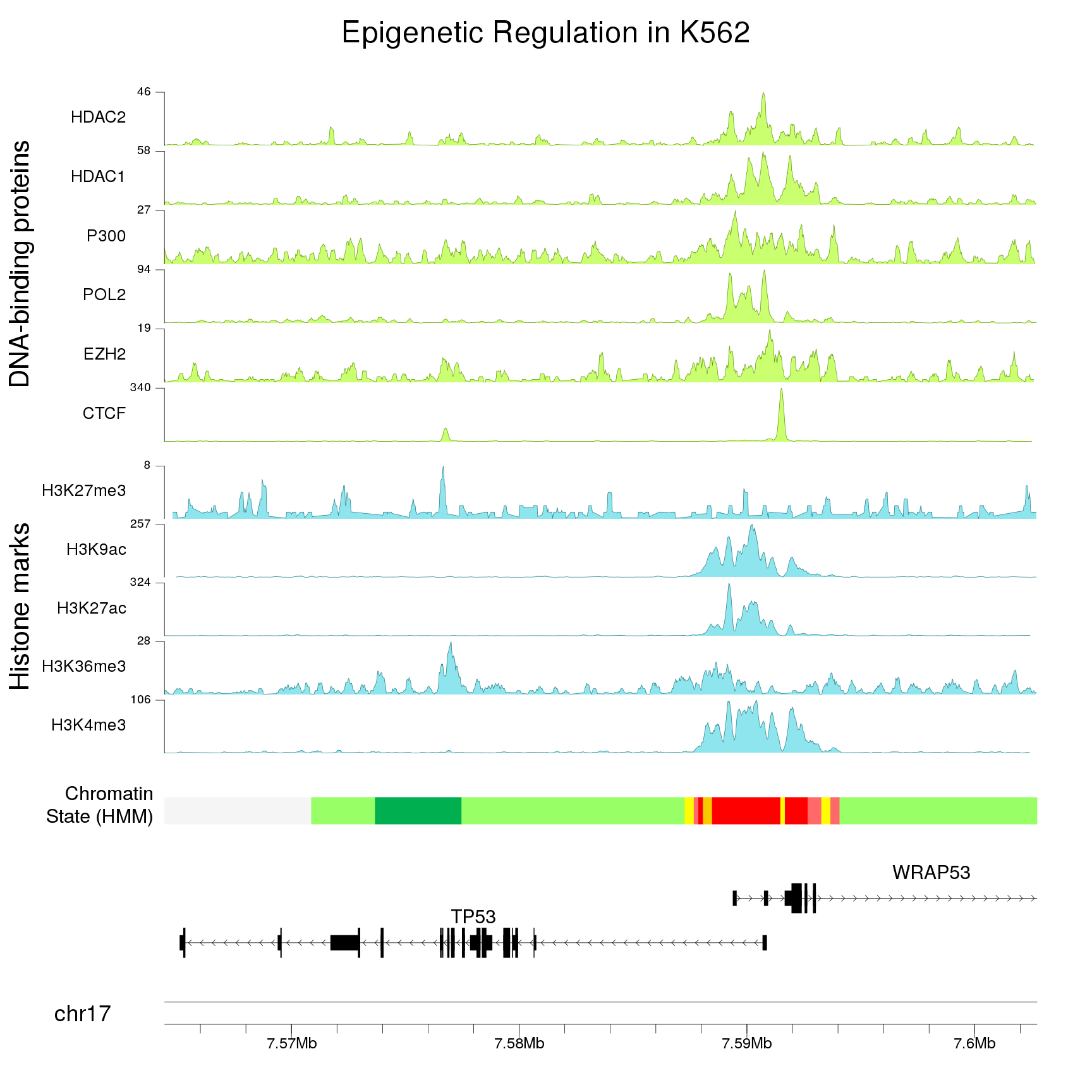
Have you checked Gviz tutorial?
https://www.bioconductor.org/packages/devel/bioc/vignettes/Gviz/inst/doc/Gviz.html#8_bioconductor_integration_and_file_support
I added a few line of my file to the post, hopefully that helps