Hello, I have a plenty of scaffold and contig names which are non-numeric like Scaffold1 (or any other name. That's why I cannot replace them with number since I have names like Fvb1 etc) instead of "1".
So, can somebody offer a package in R to get Manhattan plots with non-numeric chromosome identifiers? Also, the x axis should be adjustable and not limited to fixed chr number.
Thanks!

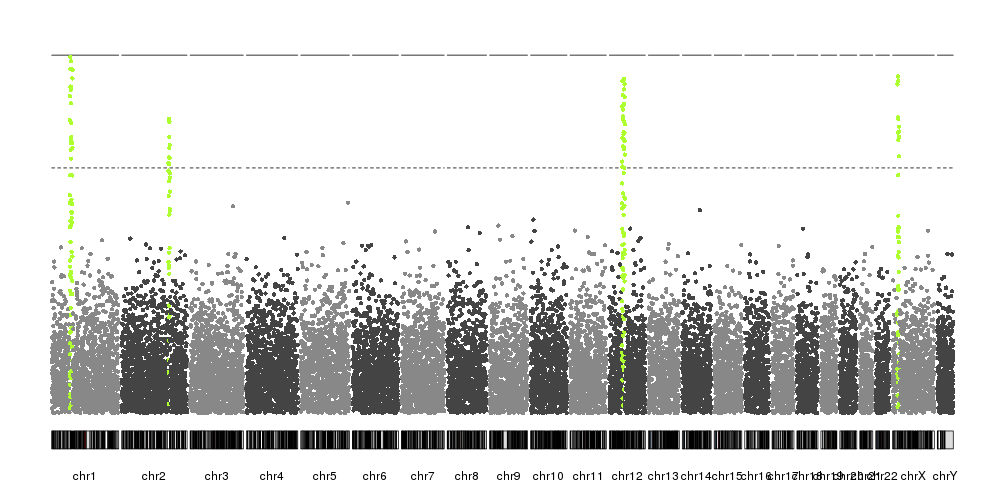
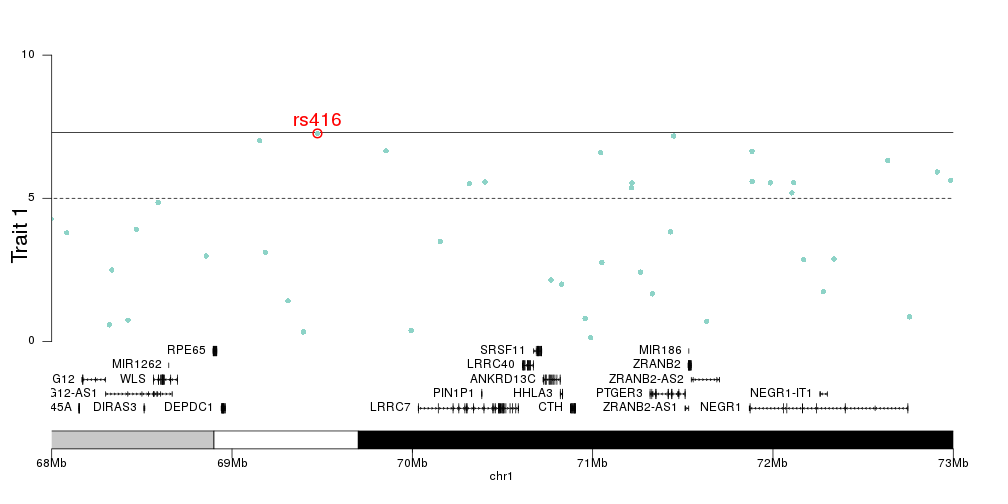

qqman does not seem feasible since I have ~ thousands unique scaffold names.. But if you still know a better way to use qqman for so many chr's, I am open any suggestions.