These tools and resources have been published in Briefings in Bioinformatics: https://academic.oup.com/bib/advance-article/doi/10.1093/bib/bbaa231/5917007
fastv is an ultra-fast tool for identification of SARS-CoV-2 and other microbes from sequencing data. It detects microbial sequences from FASTQ data, generates JSON reports and visualizes the result in HTML reports. This tool can be used to detect viral infectious diseases, like COVID-19. This tool supports both short reads (Illumina, BGI, etc.) and long reads (ONT, PacBio, etc.)
fastv is an OpenGene project: https://github.com/OpenGene/fastv
how it works?
fastv accepts FASTQ files as input, and then:
- performs data QC and quality filtering as
fastpdoes (cut adapters, remove low quality reads, correct wrong bases). - scans the clean data to collect the sequences that containing any unique KMER, or can be mapped to any reference microbial genomes.
- make statistics, visualize the result in HTML format, and output the results in JSON format.
- output the on-target sequencing reads so that they can be analyzed by downstream tools.
understand the input
fastv accepts following files as input:
- (required) the
FASTQfile to be scanned, can be single-end (-i) or paired-end (-iand-I), can be short reads (Illumina, MGI, etc.) or long reads (PacBio, ONT, etc.) - (optional) the
Genomesfile: a FASTA file containing one or many reference genomes of the target microorganism (-g). - (optional) the
KMERfile: a FASTA file containing the UNIQUE KMER of the target microbial genomes (-k). - (optional) the
KMER Collectionfile: a FASTA containing the unique KMERs of many microorganisms (-c). See an example: http://opengene.org/kmer_collection.fasta
If none of (KMER, KMER Collection, Genomes) files is specified, fastv will try to load the SARS-CoV-2 Genomes/KMER files in the data folder to detect SARS-CoV-2
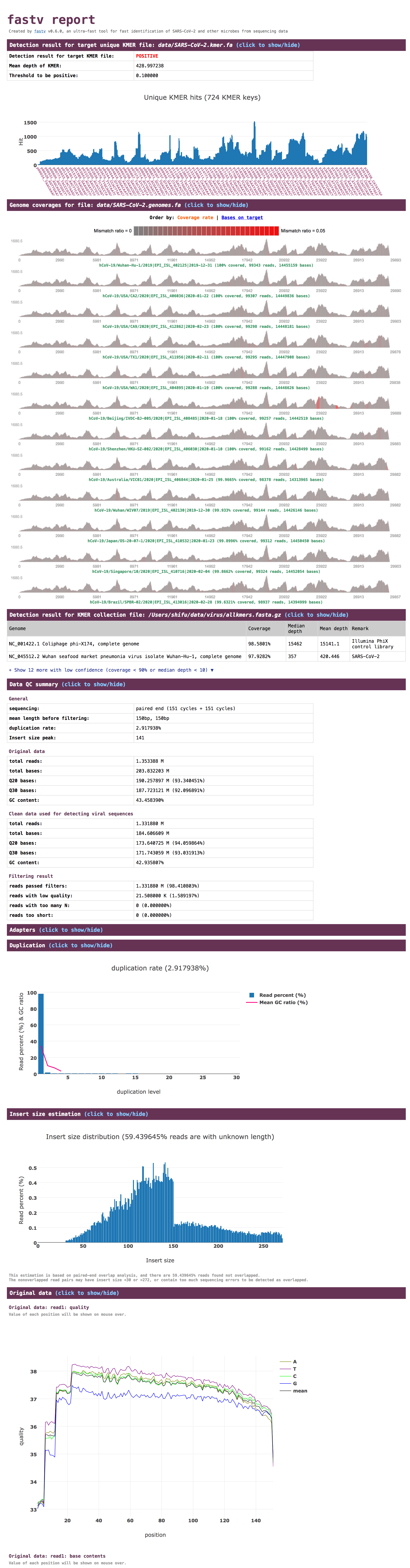
take a quick glance of the informative report
- Sample HTML report (Illumina): http://opengene.org/fastv/fastv.html
- Sample HTML report (ONT): http://opengene.org/fastv/ont.html
- Sample JSON report: http://opengene.org/fastv/fastv.json
try fastv to generate above reports
- FASTQ file for testing: http://opengene.org/fastv/testdata.fq.gz
- Command for testing:
./fastv -i testdata.fq.gz
quick examples
Single-end data
./fastv -i testdata.fq.gz
Paired-end data
./fastv -i R1.fq.gz -I R2.fq.gz
You can download KMER files and Genome files of viruses from http://opengene.org/uniquekmer/virus/index.html. This is generated by extracting unique KMERs for all genomes in a big FASTA (http://opengene.org/viral.genomic.fasta), which contains all NCBI complete RefSeq release of viral sequences that can be found from https://ftp.ncbi.nlm.nih.gov/refseq/release/viral/. The KMERs that can be mapped to human reference genome (GRCh38) with edit_distance <= 3 have already been filtered out.
You can download the KMER collection file for viral genomes from: http://opengene.org/virus.kmer_collection.fasta.gz
If you want to generate your own unique KMER files and KMER collection files, please use UniqueKMER: https://github.com/OpenGene/UniqueKMER
screenshot
For more information, go: https://github.com/OpenGene/fastv

