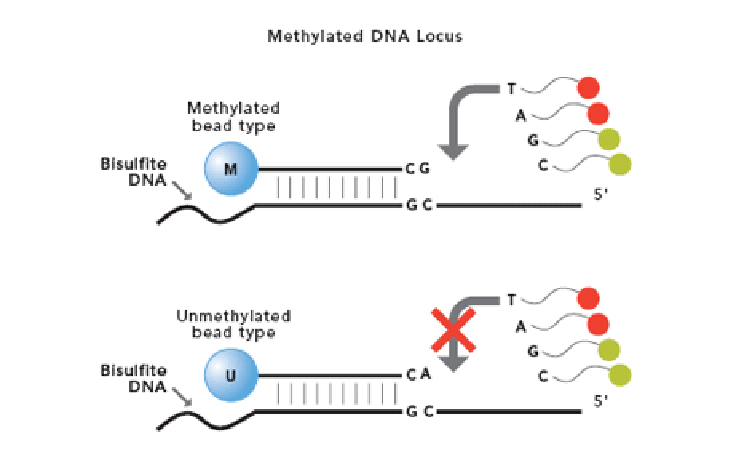
Hello everyone, I'd be very grateful if someone was to help me understand a certain step in the Illumina Infinium 450K method.
From what I've understod there are two probes, one matching the methylated DNA strand (C) and one matching the unmethylated DNA strand (T). The probes and the DNA strands will hybridize. After this I sort of get lost. One explination says the following;
Allele-specific primer annealing is followed by single-base extension using DNP- and Biotin-labeled ddNTPs. Both bead types for the same CpG locus will incorporate the same type of labeled nucleotide, determined by the base preceding the interrogated “C” in the CpG locus, and therefore will be detected in the same color channel (Figure 1). After extension, the array is fluorescently stained, scanned, and the intensities of the unmethylated and methylated bead types measured.
However I do not understand why the ddNTPs are so important. ddNTPs themsleves will extend both the methylated and the nonmetylated strands. How will we be able to diffirentiate between the methylated and nonmetylated strands if the fluroescent antibodies target the ddNTPs and not the probes themselves?
I get especially lost when they say "the base preceding the interrogated "C" in the CpG locus and therefore will be detected in the same colour channel".
I feel like I'm missing something very important. Thank you for taking your time to read this!
Best regards, A confused student.