Immunarch (https://immunarch.com), an R package for fast and painless single-cell/bulk exploration of T-cell and antibody repertoires, is finally on CRAN.
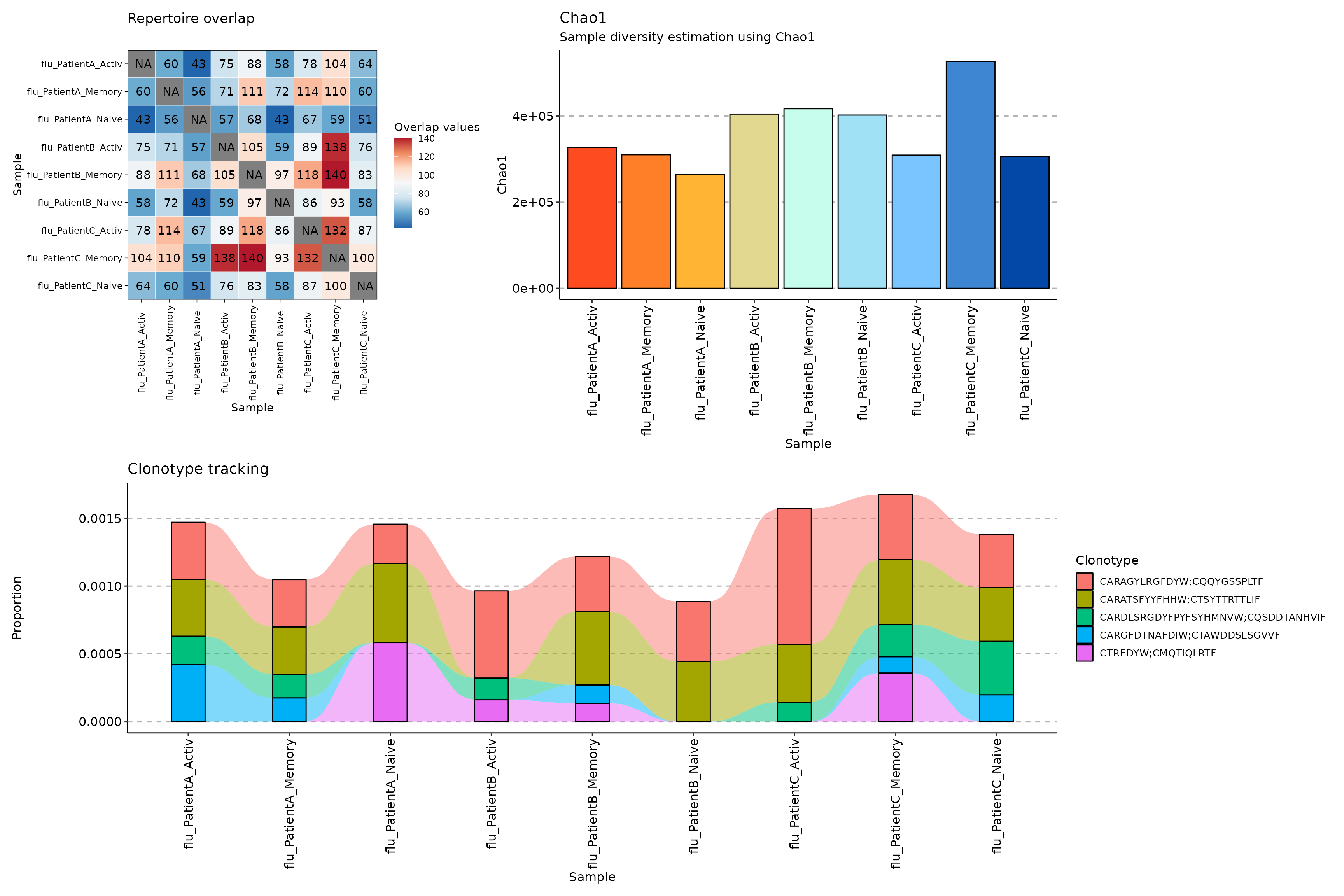
Immunarch is created to improve on the widely popular tcR package, and it makes the AIRR data exploration and visualisation simple for beginners and rapidly implementable for seasoned researchers. E.g., the following graph can be created in 1-4 clear lines of reproducible code:
Lightning-fast start:
- installation:
install.packages("immunarch") - loading the example data:
library(immunarch); data(immdata) - loading your own data: https://immunarch.com/articles/v2_data.html
- single-cell analysis: https://immunarch.com/articles/web_only/v21_singlecell.html
Our next major community & development milestones:
- a comprehensive tutorial for beginners on how to get from T-cell immune repertoire FASTQ data to publication-ready plots using immunarch. It will use COVID-19 data from the largest up-to-date collection of COVID-19 datasets: https://github.com/immunomind/covid19
- full interoperability with Seurat for streamlined single-cell paired chain data exploration
- a detailed tutorial on how to speed up immune repertoire pipelines by 5-10 times
- advanced BCR support, including parsing and mutation analysis for CDR1-2 sequences
All immunarch news and updates will be in this thread for your convenience. If you prefer other means of monitoring, please feel free to follow our Twitter: https://twitter.com/immunomind
Our team will be glad to answer your questions, promptly fix a bug or implement a feature, just create a ticket here: https://github.com/immunomind/immunarch/issues GitHub is our major hub for the communication and it is monitored hourly.
Your suggestions, GitHub stars and code contributions are warmly welcome! To start, visit our GitHub page: https://github.com/immunomind/immunarch
A detailed documentation is here: https://immunarch.com/

