Hi everybody. I am using the online version of STRING (https://string-db.org) to build a PPI network using DEGs from RNAseq analysis. In the settings, I checked the "molecular action" box to visualize the predicted mode of action for the interaction between different nodes. But when I export the .tsv file from the site to load the network on cytoscape, this information is not present.
I checked all the files that can be exported and there is no trace of "molecular action". Do you know how can I export this type of info to load it into other software like cytoscape?
Or is there any other way to do this? Or other software/tools/websites? The STRING app in cytoscape do not allow to visualize the "molecular action".
Thank you for your help!
ps here's a picture of a network as an example of what I can see on STRING (with "molecular action" turned on). The different edges that you can see are what I want to export (e.g. green arrow for activation, red line for inhibition, etc).
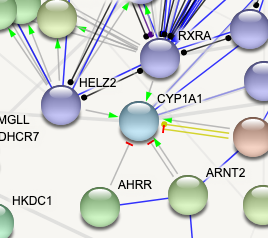


Thank you for your answer! I am trying to export the network in cytoscape; is there a way to then import this information (molecular action) into the network inside cytoscape?
File → Import → Table from File…. and selecting appropriate options or if this doesn't work, just pre-process the file, for example replacing the gene IDs to match those you already have. To reproduce the STRING visualization, you'll also need to play with styles.