Entering edit mode
4.3 years ago
alireza.khodadadi.j
▴
70
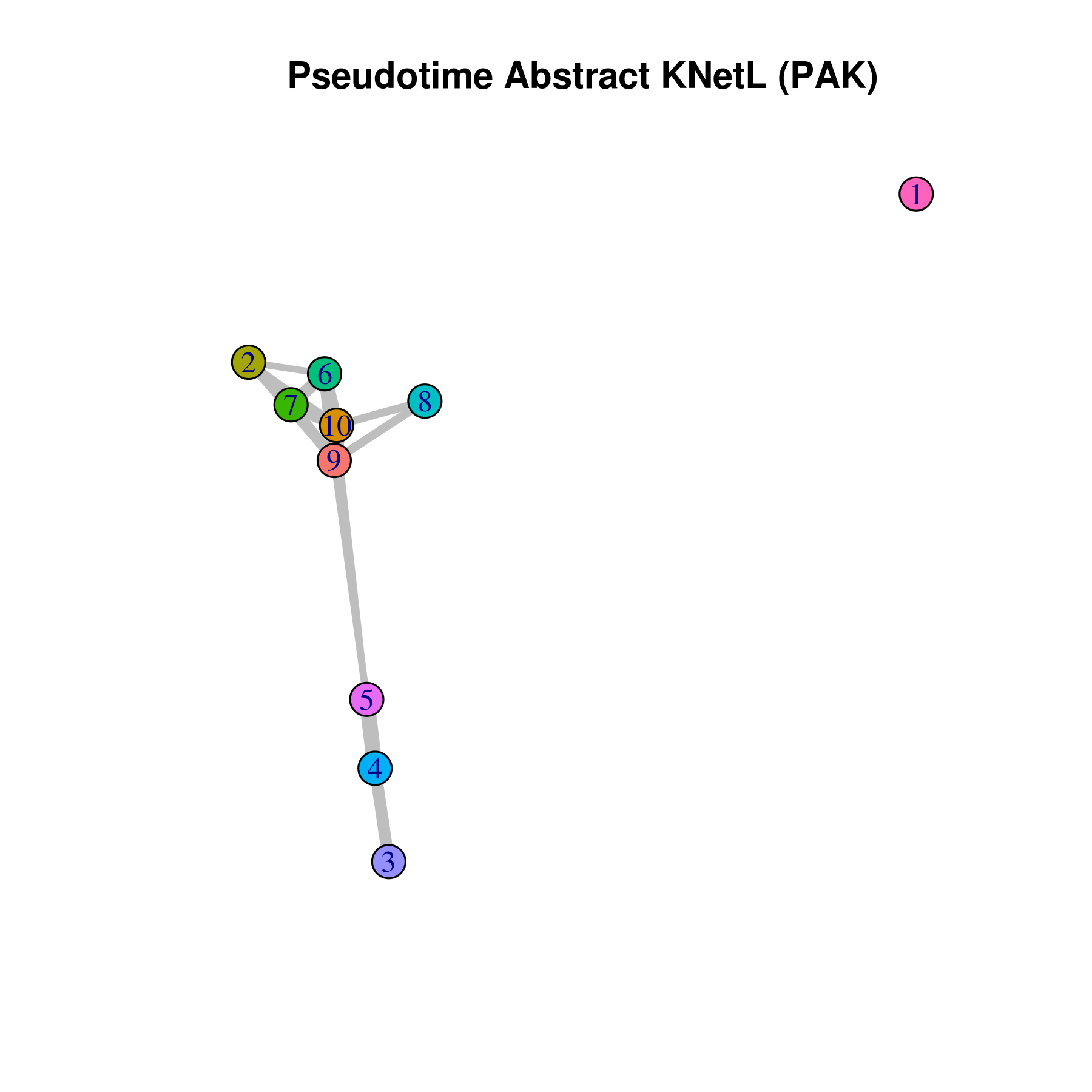
Pseudotime Abstract KNetL map (PAK map) helps finding the relationships and the true distance between clusters in each condition. In PAK maps each node represents a cluster and the length and the thickness of the links (edges) represent distances between each cluster. The shorter and thicker the link the more related (similar) the cell communities.
PAK map is available through iCellR (v1.5.5) package:
https://github.com/rezakj/iCellR (GitHub)
https://CRAN.R-project.org/package=iCellR (CRAN)
How to run an make PAK maps using iCellR:
pseudotime.knetl(my.obj,interactive = F,cluster.membership = F,conds.to.plot = NULL)