Entering edit mode
13.0 years ago
garyfox111
•
0
Hi I'm examining a genome from Ensembl, and its pretty simple the work out the number of exons per gene from that data, but working out the introns from that has me a bit confused, Is it simply the number of exons minus 1? or is it the same? Thanks

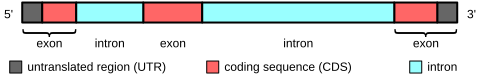

The only part you should be careful about is making the distinction between the number of exons in a transcript (ie, one isoform of a gene) and the gene itself. With alternative splicing, these numbers are not always equal.
This diagram may not be complete, as UTRs can also have introns.
Yes. +1 for Eric's comment. If you want the number of introns for a "gene" (locus) you will need to add together the (unique) introns on a transcript-by-transcript basis. Most genes have multiple transcripts and there are many ways that transcripts can differ such that some introns will be shared between transcripts, some will be unique and some will be overlapping. Given the latter, you will need to compare coordinates to determine uniqueness of introns.