Entering edit mode
4.7 years ago
pthom010
▴
40
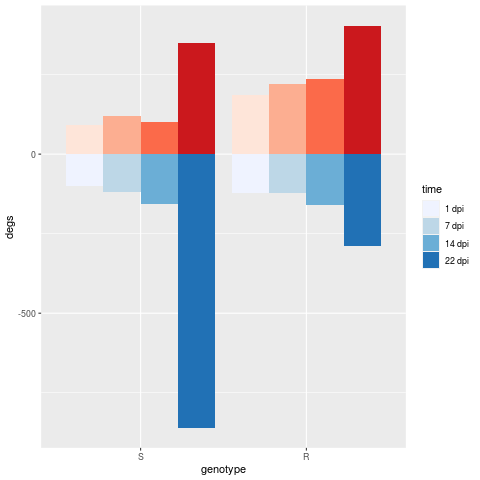
I have a plot that I have made counting the number of genes over a period of time in two samples. I would like to plot both sets of numbers in a waterfall chart. I am able to get the chart but I have difficulty getting both legends to show up. I have the following tables:
deg.up
genotype time degs
1 S 1 dpi 91
2 S 7 dpi 121
3 S 14 dpi 102
4 S 22 dpi 349
9 R 1 dpi 186
10 R 7 dpi 220
11 R 14 dpi 236
12 R 22 dpi 404
deg.dn
genotype time degs
5 S 1 dpi -99
6 S 7 dpi -120
7 S 14 dpi -157
8 S 22 dpi -860
13 R 1 dpi -121
14 R 7 dpi -122
15 R 14 dpi -161
16 R 22 dpi -288
I am using the following code to make the plot:
deg.dn$degs=deg.dn$degs*-1
deg.up$time=as.character(deg.up$time)
deg.dn$time=as.character(deg.dn$time)
deg.up$time = factor(deg.up$time, levels=unique(deg.up$time))
deg.dn$time = factor(deg.dn$time, levels=unique(deg.dn$time))
deg.up$genotype=as.character(deg.up$genotype)
deg.dn$genotype=as.character(deg.dn$genotype)
deg.dn$genotype = factor(deg.dn$genotype, levels=unique(deg.dn$genotype))
deg.up$genotype = factor(deg.up$genotype, levels=unique(deg.up$genotype))
breaksup = levels(deg.up$time)
breaksdn = levels(deg.dn$time)
brewup = c("#FEE5D9", "#FCAE91", "#FB6A4A", "#CB181D")
brewdn = c("#EFF3FF", "#BDD7E7", "#6BAED6", "#2171B5")
ggplot() +
geom_bar(data = deg.up, aes(x=genotype, y=degs, fill=time), stat="identity", position="dodge") +
scale_fill_manual(values=brewup, breaks=breaksup) +
new_scale_fill() +
geom_bar(data = deg.dn, aes(x=genotype, y=degs, fill=time), stat="identity", position="dodge") +
scale_fill_manual(values=brewdn, breaks=breaksdn)
Would anybody be able to help me get both legends next to each other in the final plot? Thank you.!

