Hi mag,
Another option would be to use karyoploteR to create your Manhattan plot. The plot will be a bit different since karyoploteR will plot each SNP in its correct position on the genome, by default on hg19 (note that karyoploteR can work with any genome, though).
To use it, you'll first need to convert your data into a GenomicRanges object, and change the chromosome names to UCSC style.
snps.gr <- toGRanges(dd[,c(4,5,5,6,1,2,3)])
seqlevelsStylesnps.gr) <- "UCSC"
And you are good to go. We'll create a plot with plotKaryotype, then add an axis with kpAxis and finally use kpPlotManhattan to actually plot the data. To give each point a color according to its group, we'll use the colByCategory function, and that's it.
kp <- plotKaryotype(plot.type=4)
kpAxis(kp, ymin = 0, ymax = 10)
kpPlotManhattan(kp, data = snps.gr, pval = snps.gr$`-log10_Pval`, logp = FALSE, ymin=0, ymax=10,
points.col=colByCategory(snps$group, colors = c("red", "blue")))
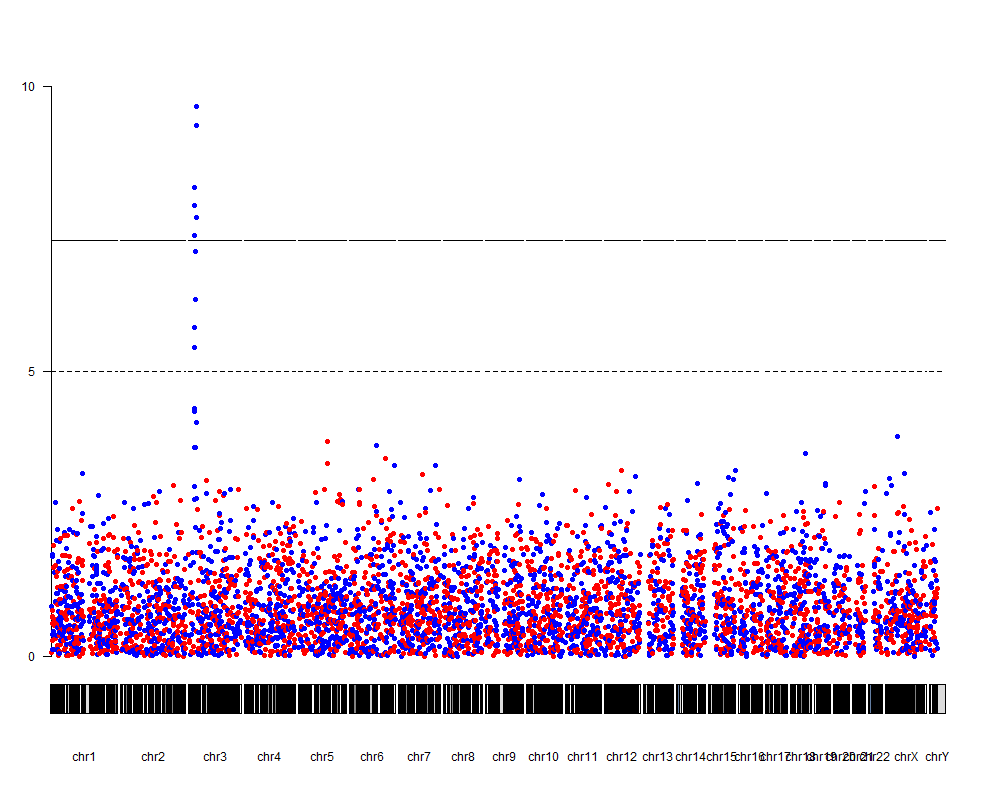
With a few additional tweaks we can improve the plot and annotate the top snps, for example.
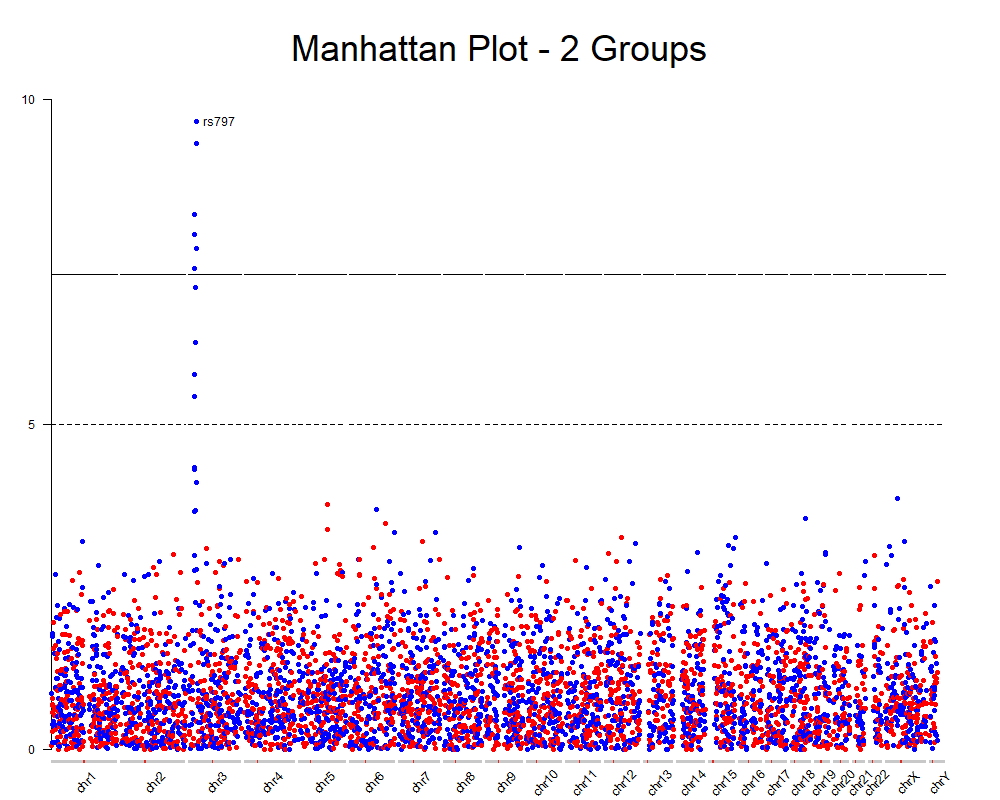
You can find the complete code for this on github and all the instructions on how to create Manhattan plots with karyoploteR and how to add additional data into a Manhattan plot on the karyoploteR tutorial.
Hope this helps




Please see How to add images to a Biostars post to add your images properly. You need the direct link to the image or the HTML embed code, not the link to the webpage that has the image embedded (which is what you have used here)
str(my_data)orhead(my_data)?see if this is what you want: