Entering edit mode
4.4 years ago
dimitrischat
▴
210
Hi all. Dont know how to solve this, so here it goes. trying to do an rna-seq analysis on solanum lycopersicum. I am at the step of htseq-count so i got the gff from this website: https://solgenomics.net/organism/Solanum_lycopersicum/genome/ ->> ITAG4.0. I used to do rna-seqs on humans so in the hg19.gtf file there were "gene_id" therefore i could use the option " -i gene_id " when executing htseq-count command. On this gff (a portion of the gff file):
SL4.0ch00 maker_ITAG gene 417592 418482 . + . ID=gene:Solyc00g500004.1;Alias=Solyc00g500004;Name=Solyc00g500004.1;length=890
SL4.0ch00 maker_ITAG mRNA 417592 418482 . + . ID=mRNA:Solyc00g500004.1.1;Parent=gene:Solyc00g500004.1;Name=Solyc00g500004.1.1;Note=Unknown protein;_AED=0.47;_QI=0|0|0.5|1|0|0|2|524|98;_eAED=0.56
SL4.0ch00 maker_ITAG exon 417592 417723 . + . ID=exon:Solyc00g500004.1.1.1;Parent=mRNA:Solyc00g500004.1.1
SL4.0ch00 maker_ITAG CDS 417592 417723 . + 0 ID=CDS:Solyc00g500004.1.1.1;Parent=mRNA:Solyc00g500004.1.1
SL4.0ch00 maker_ITAG CDS 417794 417958 . + 0 ID=CDS:Solyc00g500004.1.1.2;Parent=mRNA:Solyc00g500004.1.1
SL4.0ch00 maker_ITAG exon 417794 418482 . + . ID=exon:Solyc00g500004.1.1.2;Parent=mRNA:Solyc00g500004.1.1
SL4.0ch00 maker_ITAG three_prime_UTR 417959 418482 . + . ID=three_prime_UTR:Solyc00g500004.1.1.0;Parent=mRNA:Solyc00g500004.1.1
Is there an option on htseq-count i could use for the genes? Or is there a way to convert this gff file to the one like the hg19.gtf - image below
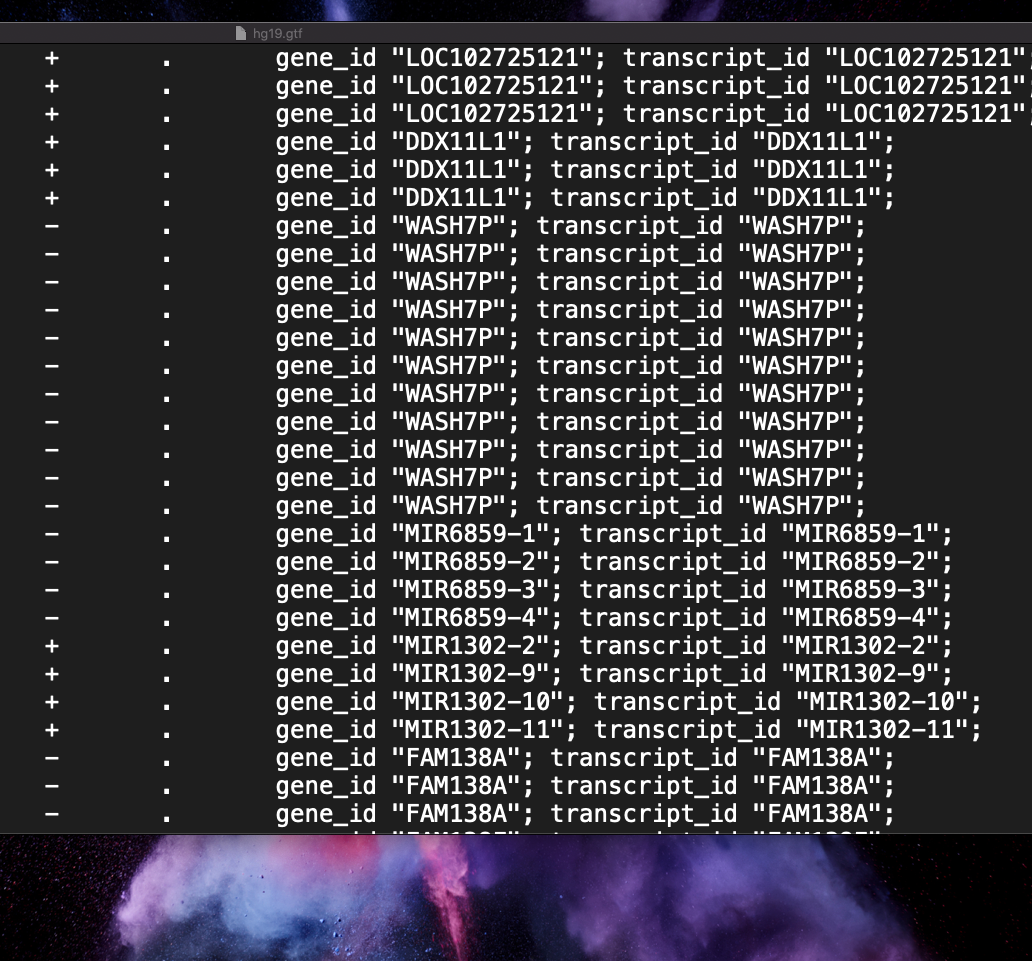


Please see How to add images to a Biostars post to add your images properly. You need the direct link to the image or the HTML embed code, not the link to the webpage that has the image embedded (which is what you have used here)