With KEGG, we can use the tool Bioconductor to plot our pathways with the logfold changes for our datasets, using R. Like this:
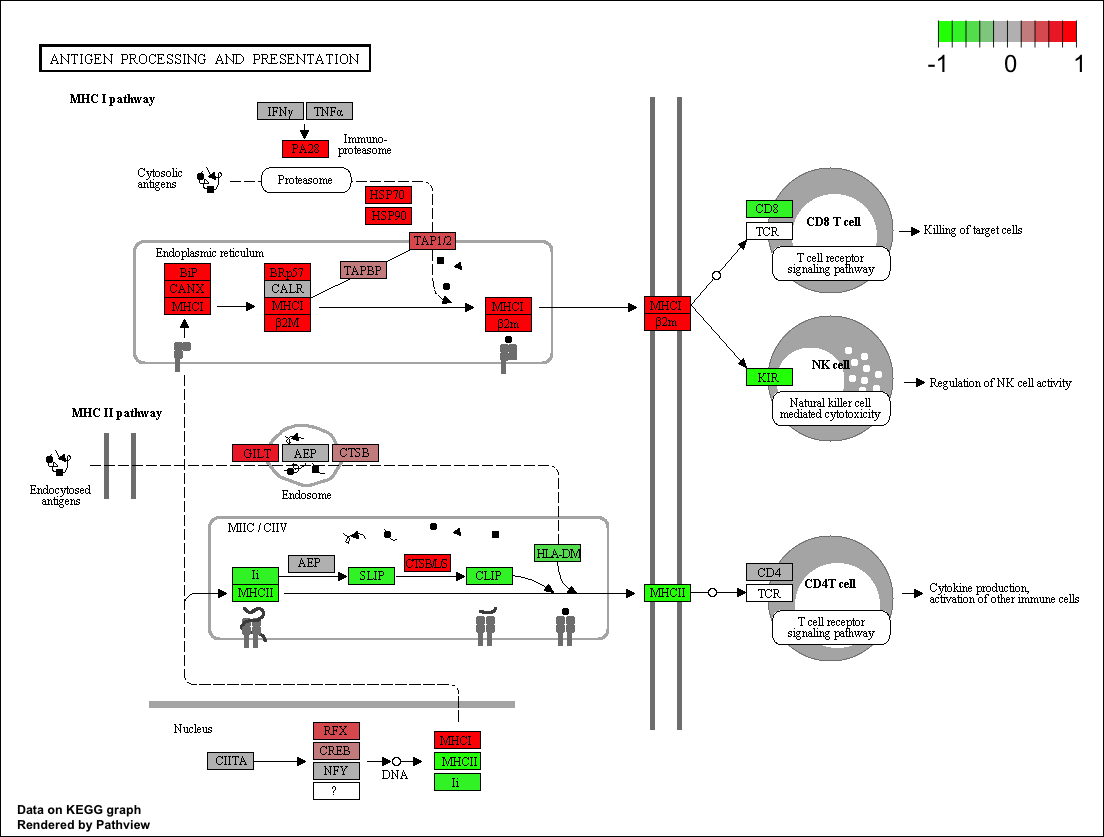 This can also be done with WikiPathways and Cytoscape, by downloading the images and uploading the fold changes into cytoscape via R tools such as RCy3.
This can also be done with WikiPathways and Cytoscape, by downloading the images and uploading the fold changes into cytoscape via R tools such as RCy3.
Can the same be done with Reactome? I preferrably want the pathways in the original representation as shown on the svg file on the reactome website. Like this:
But with log fold changes. There is a way to do this in ReactomePA, but it shows the result in a dissociated network diagram that is hard to make sense out of.






Hi, Gabriel!
Any chance you found out how to do this?
Please do not add comments as answers - use the "Add Comment" or "Add Reply" buttons as necessary to help keep things organized.
Additionally, piggybacking on old questions is rarely an effective method to get help, though we do appreciate you searching for them. If you have a question and cannot find the answer, please post a new question with sufficient detail to understand your issue(s) and goal(s), ideally with a reproducible example if possible.
Sorry, that was unintentional, but completely my bad! Thanks for fixing it.
Since there were no solutions to this here, I was not sure how else to contact Gabriel to ask if maybe they had figured this out. Is commenting on an un-answered thread to get it back on the 'latest' queue and potentially getting clarifications from the OP worse than re-posting a similar/same question again? I didn't know that, it felt more intuitive to expand on something already written to have it all in one place instead of another separate thread, but I will keep this in mind, thanks.
This post is a year and a half old and Gabriel is not an active user. Regardless, your question buried in a comment is a lot less likely to get attention from other users than a new one. You can even link this question, mention that it got no attention, and provide details specific to your situation, etc. That shows you made the effort to search the site first.
However, a question as straightforward as this would probably have an answer if there was one easily found (and I'm sure you've done your fair share of googling as well). Feel free to open a new question, but I wouldn't get your hopes up too high.
Thanks for the clarification and the additional tips, Jared!