The "invgamma" setting in MrBayes sets a gamma-distributed rate variation across sites and a proportion of invariable sites. The gamma distribution is approximated by several discrete rate categories.
According to Felsenstein 1981 (Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach), the site likelihood is computed from the root-node conditional likelihoods by $L=\sum_{s_0} \pi_{s_0} L_{s_0}^{(0)}$. However, when the invgamma setting is used in MrBayes, there will be a root-node conditional likelihood for each discrete rate category.
What effect do the discrete rates have on computing the site likelihood from the root-node conditional likelihoods?
What effect does the "proportion of invariable sites" have on the computation?
In particular, I'm looking for some documentation that answers these questions in detail.

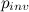 be the proportion of invariant sites, n the number of gamma categories and
be the proportion of invariant sites, n the number of gamma categories and 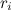 the rate corresponding to the
the rate corresponding to the 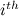 category of the discrete gamma model (as determined by the actual gamma shape parameter). Let
category of the discrete gamma model (as determined by the actual gamma shape parameter). Let 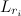 be the root-node conditional likelihood calculated with a given fixed rate
be the root-node conditional likelihood calculated with a given fixed rate ![L(p_{inv}, r_{1}, r_{2},\ldots,r_{n}) = p_{inv} L(0) + (1 - p_{inv}) [\frac{1}{n} \sum_{i=1}^{n} \L(r_i)]](http://mathurl.com/2u4tv9m.png)

This might be an implementation detail that the developers might know more about.