Dear users, Since many days I'm struggling with a technical problem for which I was not able to find any clear answer given that I'm not familiar with the automatic manipulation of phylogenetic trees. let's consider the following phylogeny tree :
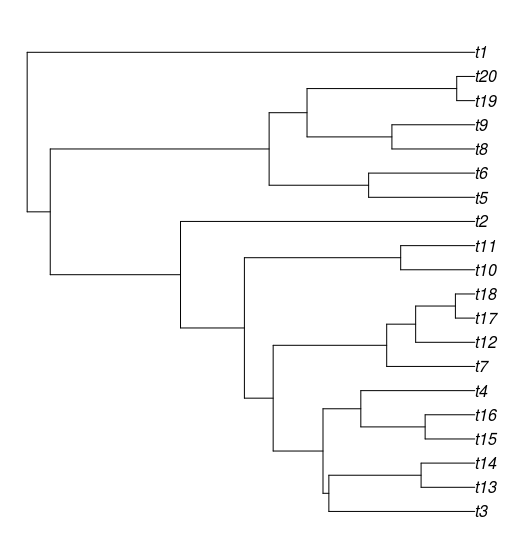
It there any automatic way (R package or any other langue) to get the closest taxa names of a particular target taxa. For example the closest species of t3 will be : t14 and t13. for t7 it will be : t12,t17,t18 and for t10 this will be t11. It is also possible to add a threshold using bootstrap values ?
Thank you so much for your help !
Cheers


Dear a.zielezinski , thanks for your answer. I tried your code and it works ! however the closest leaves to weasel in your example is cat and monkey. However what I wanted to get based on your example is the sister clade that contain dog, bear, raccoon, seal and sea_lion not the outgroups (cat and monkey)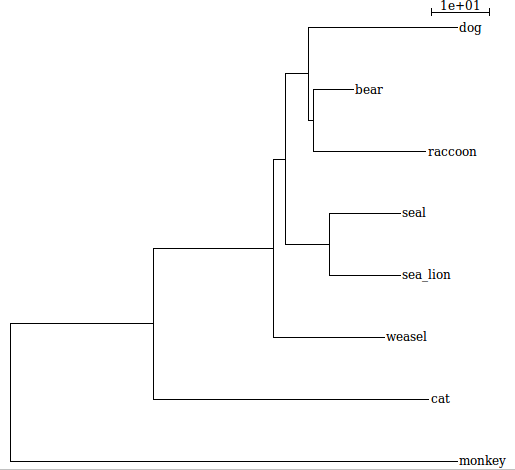
Do you know if it is possible to consider sister group only if the bootstrap value is bigger than a threshold
Thanks again for your help !
Hi mchimich, I just updated my answer. Hope it helps.
Thank you so much !! now everything is working perfectly ! appreciated