I'm trying to draw a Sankey plot. But while plotting I'm getting some issues as some target and source nodes are missing and also the edges/links are not as same color as source node.
My Sankey Code:
library(tidyverse)
library(viridis)
library(patchwork)
library(hrbrthemes)
library(circlize)
library(networkD3)
library(readxl)
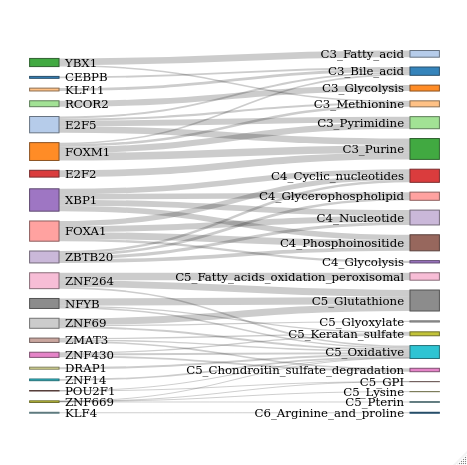
DF <- read.delim(file = "FOR_SANKEY.txt")
# Make a list of total number of
nodes <- data.frame(name=c(as.character(DF$source), as.character(DF$target)) %>% unique())
# Assign IDs to source and target
DF$IDsource=match(DF$source, nodes$name)-1
DF$IDtarget=match(DF$target, nodes$name)-1
# Assign color scale
ColourScal ='d3.scaleOrdinal(["#FF5733","#33DBFF","#FF8733","#FF9F33","#FFF033", "#DFFF33", "#FF3351", "#33FFE1", "#EB33FF", "#47FF33", "#9733FF", "#338CFF", "#6BFF33", "#C733FF", "#9D33FF", "#95FF33", "#33ECFF", "#FF4633", "#DAFF33", "#5833FF"])'
# Make the Network
sankeyNetwork(Links = DF, Nodes = nodes, Source = "IDsource", Target = "IDtarget", Value = "value", NodeID = "name", sinksRight=FALSE, colourScale=ColourScal, nodeWidth=20, fontSize=12, iterations = 0)
My input Data:
source target value IDsource IDtarget
CEBPB C3_ Bile acid -0.389361351 0 20
E2F5 C3_ Bile acid -0.395798119 3 20
FOXM1 C3_ Bile acid -0.431892661 5 20
KLF11 C3_ Bile acid -0.245382431 6 20
YBX1 C3_Fatty acid 0.352803344 12 21
RCOR2 C3_Glycolysis 0.234987627 10 22
E2F5 C3_Methionine -0.372301522 3 23
FOXM1 C3_Methionine -0.302098443 5 23
YBX1 C3_Methionine -0.45913054 12 23
E2F2 C3_Purine 0.376737908 2 24
E2F5 C3_Purine 0.325288339 3 24
FOXM1 C3_Purine 0.48464042 5 24
E2F5 C3_Pyrimidine 0.230968129 3 25
FOXM1 C3_Pyrimidine 0.27487664 5 25
FOXA1 C4_Cyclic nucleotides 0.124340069 4 26
XBP1 C4_Cyclic nucleotides 0.165542857 11 26
ZBTB20 C4_Cyclic nucleotides -0.308870128 13 26
XBP1 C4_Glycerophospholipid 0.166331418 11 27
ZBTB20 C4_Glycerophospholipid -0.272486646 13 27
FOXA1 C4_Glycolysis -0.328322849 4 28
FOXA1 C4_Nucleotide 0.220425799 4 29
XBP1 C4_Nucleotide 0.227317275 11 29
ZBTB20 C4_Nucleotide -0.196174304 13 29
FOXA1 C4_Phosphoinositide 0.38027914 4 30
XBP1 C4_Phosphoinositide 0.174345152 11 30
ZBTB20 C4_Phosphoinositide -0.115285479 13 30
POU2F1 C5_ GPI -0.659795918 9 31
ZMAT3 C5_Chondroitin sulfate degradation -0.447244898 14 32
ZNF430 C5_Chondroitin sulfate degradation -0.395714286 17 32
ZNF264 C5_Fatty acids oxidation peroxisomal 0.469450751 16 33
NFYB C5_Glutathione 0.444143983 8 34
ZNF264 C5_Glutathione 0.381897498 16 34
ZNF69 C5_Glutathione 0.385724126 19 34
ZNF69 C5_Glyoxylate -0.500625016 19 35
ZNF669 C5_GPI -0.690306122 18 36
NFYB C5_Keratan sulfate -0.495918367 8 37
ZMAT3 C5_Keratan sulfate -0.479285714 14 37
ZNF430 C5_Keratan sulfate -0.482755102 17 37
ZNF669 C5_Lysine -0.659795918 18 38
DRAP1 C5_Oxidative -0.375886525 1 39
NFYB C5_Oxidative -0.458163265 8 39
POU2F1 C5_Oxidative -0.636734694 9 39
ZMAT3 C5_Oxidative -0.46622449 14 39
ZNF14 C5_Oxidative -0.424081633 15 39
ZNF264 C5_Oxidative -0.439489796 16 39
ZNF430 C5_Oxidative -0.39 17 39
ZNF669 C5_Oxidative -0.614489796 18 39
ZNF69 C5_Oxidative -0.392040816 19 39
ZNF669 C5_Pterin -0.543534274 18 40
KLF4 C6_Arginine and proline -0.537148898 7 41


Thanks Dunois for your valuable help.
If an answer was helpful, you should upvote it; if the answer resolved your question, you should mark it as accepted. You can accept more than one answer if they all work.
Thank you sir for your help.