I want to map a list of KEGG terms with their corresponding names. Is it possible to get a list of all KEGG terms and their descriptions/names?
Hi.
I am not sure if you already figured it out, but, I know two easy ways of getting KO names/descriptions and use them afterwards to map their names to the respective KO IDs.
1 - The elegant way: Get title
- If you happen to have a list of KOs, you can map their names using the lower field at "KO (KEGG ORTHOLOGY) Database" page (https://www.genome.jp/kegg/ko.html); enter your IDs (separated by either new lines, spaces, or both) and click "Get title":
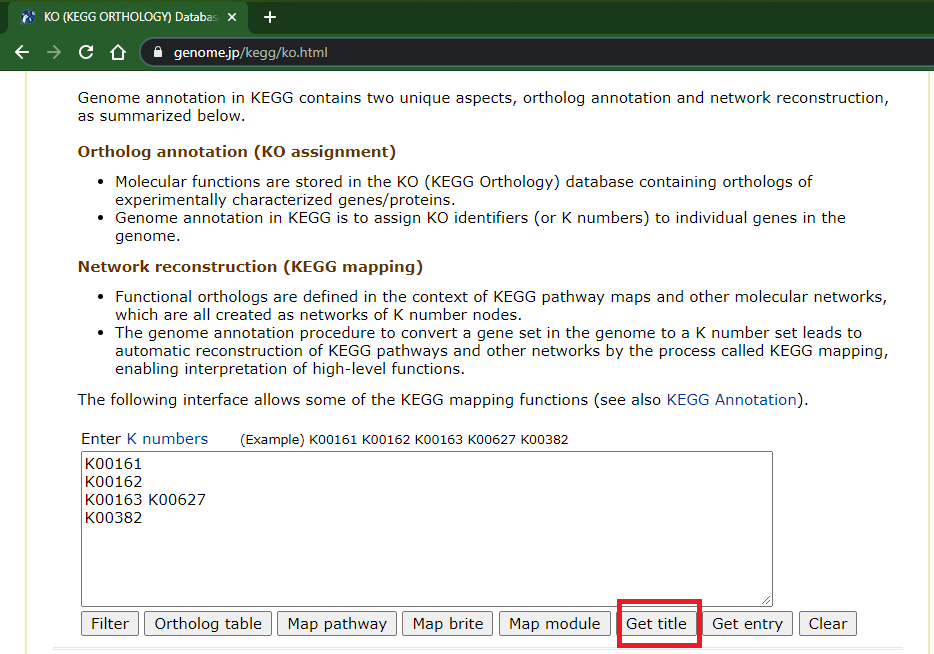
- This opens a new browser tab with the KOs description/names:
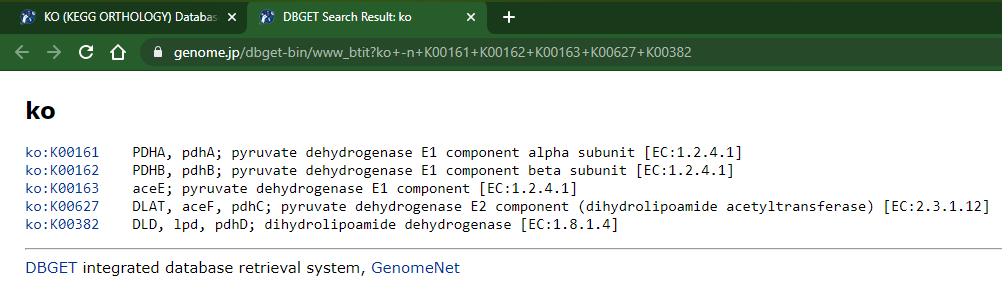
2 - The not-so-elegant way: Copy from page
If you wish to get all the descriptions/names from once, open the "KEGG Orthology (KO)" link in the "KO (KEGG ORTHOLOGY) Database" page (https://www.genome.jp/kegg/ko.html):
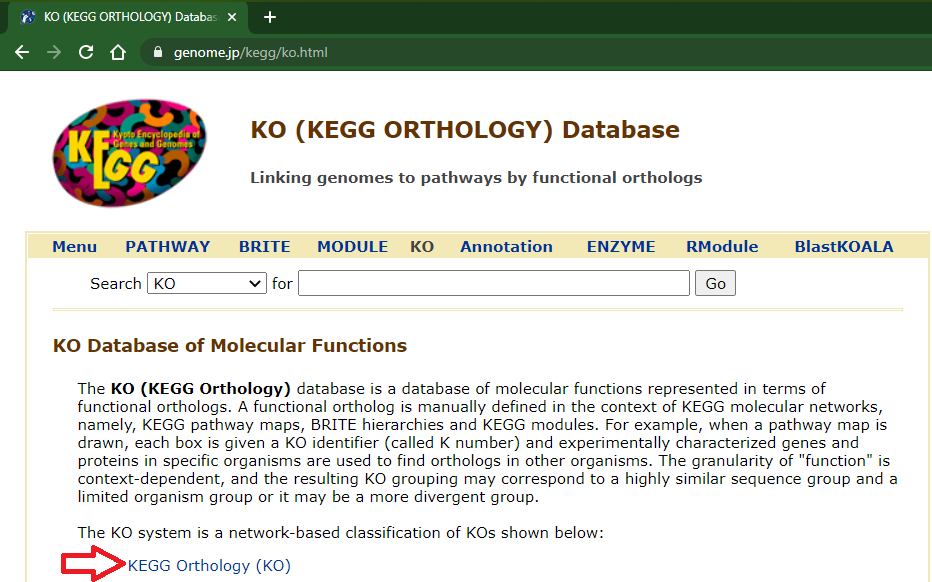
Click in the rightmost arrow at top of the list (wait the complete page load):

Then copy everything! it may lag a litle because the page is a bit heavy (55k+ lines, currently)
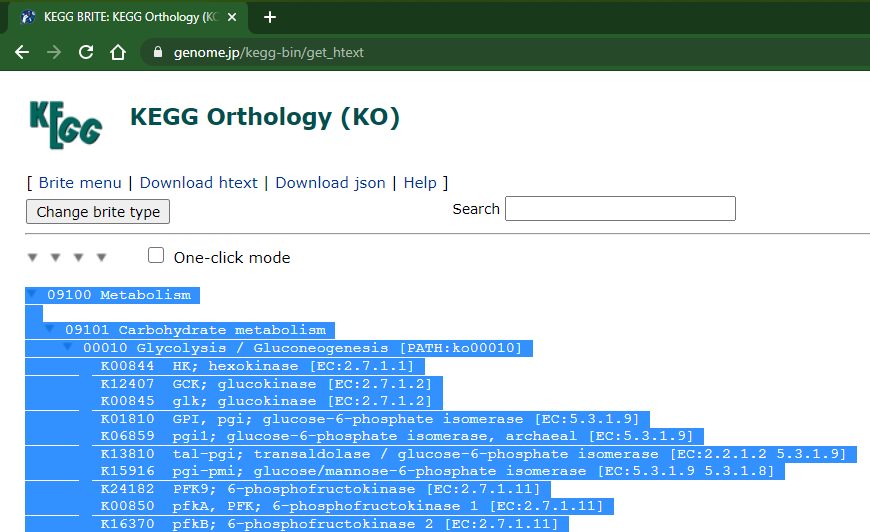
Now you can paste in a text file (in my example I used notepad++, but any other text editor should work):
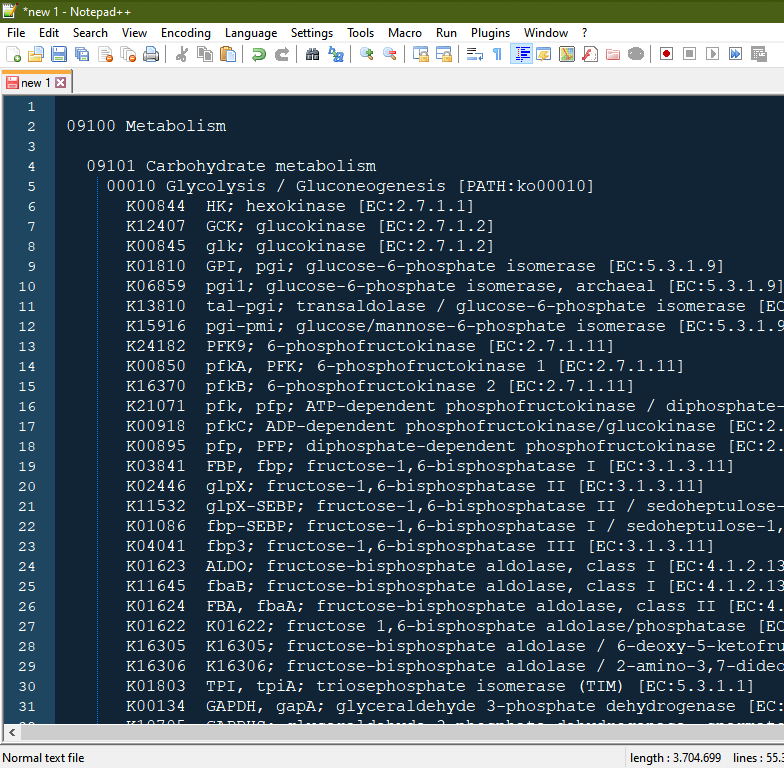
- Save as tab-delimited file or any other usefull format and get whichever description you want.
Tips
1 - Avoid using excel IF you are not REALLY sure excel won't mess up the entered data.
If you are REALLY sure it is ok, you can open the file as a speadsheet and use it as a database in formulas like VLOOKUP to map the descriptions to any KOs list you have (comment if you need me to show you how).
2 - In unix systems, you can easily get the description from the text file using grep, awk, sed, etc. This is much better if you wanna map the descriptions within a script/pipeline/program, etc.
I'm glad if this answer helps anyone. Cheers
My group prepared a very nice tool based on KEGG ( see https://dariusz-izak-doktorat.github.io/ )
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Hey Arsenal,
I just made a post about this, but since its somewhat related, do you have an idea how I can get the functional category ( different level of the heirarchy) rather than the specific gene function? I'm trying to summarize the annotations by saying for example there are 243 genes related to "Carbohydrate metabolism"