Dear Cytoscape Community,
I'm working on an interaction network with Cytoscape (Version: 3.2.8) showing connections between miRNAs and targeted genes. I'm now looking for a way to filter for the gene targets, that connect at least two miRNAs with each other to reduce the number of gene targets in my network (Step 1)
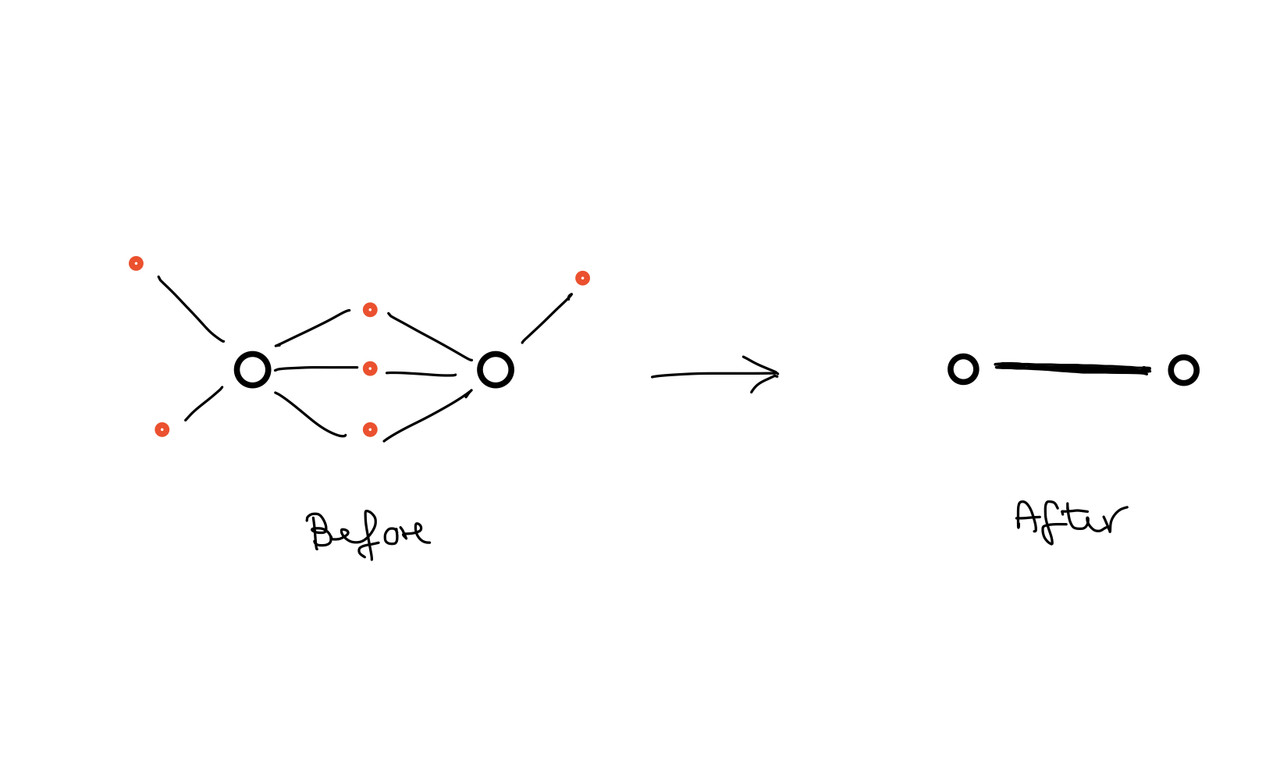
Next I would like to replace these left over connecting gene targets by edges that connect only the miRNAs. My final network should then consist only of interconnected miRNAs, where the width of the edges represents the number of connecting (but invisible) gene targets (Step 2).
Is there a possible workflow/ app to get this done? I think I could handle step 1 manually by data pre-processing. Couldn't find a solution for step two though yet. Happy for your advice. Thank you for your help!