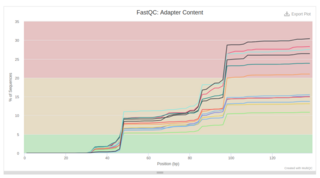
I run standard QA on SHAPEseq reads and MultiFastQC reported the presence of Nextera Transposase Seqeunce (Illumina adapter). I wanted to cut out adapters as usual with trimmomatic, but even though I specified the adapter sequence (and then double-check with minion), adapters are still visible in the post-QA report.
File with adapters:
>adapter_1_FW/1\
CCGAGCCCACGAGACCTCTCTACATCTCGTATGCCGTCTTCTGCTTG
>adapter_1_RV/2\
ACGCTGCCGACGATCGCATAAGTGTAGATCTCGGTGGTCGCCGTATCATT
>adapter_2_FW/1\
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG
>adapter_2_RV/2\
GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG
>adapter_3_FW/1\
GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAG
>adapter_3_RV/2\
GCCTTGCCAGCCCGCTCAGAGATGTGTATAAGAGACAG


Try
bbduk.shas an alternative. A guide is available.Thank you for suggestion! But the strange thing is that I run also other SHAPEseq reads with the same adapters and they were cut out correctly.
Can you post the following: (1) the trimmomatic options you used, (2) trimmomatic log file for one sample, (3) the first few lines of the file containing the adapter sequence.
Suer - please see the edited post