Dear colleagues,
We are happy to announce the release of elPrep 4.0.0, an open-source, drop-in replacement tool for GATK4/Picard/SAMtools for preparing SAM/BAM files for variant calling that produces identical results, while greatly improving computational performance. For more details, see the elprep github repository.
elPrep 4.0.0 introduces multiple new features allowing us to process the preparation steps defined by the GATK Best Practices for variant calling.
New features include:
- added base quality score recalibration (BQSR)
- added optical duplicate marking
- added metrics (MultiQC compatible)
- support for SAM File Format version 1.6
- support for FASTA and VCF files
- support for elPrep-specific elsites and elfasta formats
- split/filter/merge (sfm) mode now implemented in Go instead of Python
- added --log-path option to all tools
- various API and performance improvements
- changed license to the GNU Affero General Public License version 3 as published by the Free Software Foundation, with Additional Terms
- updated demos
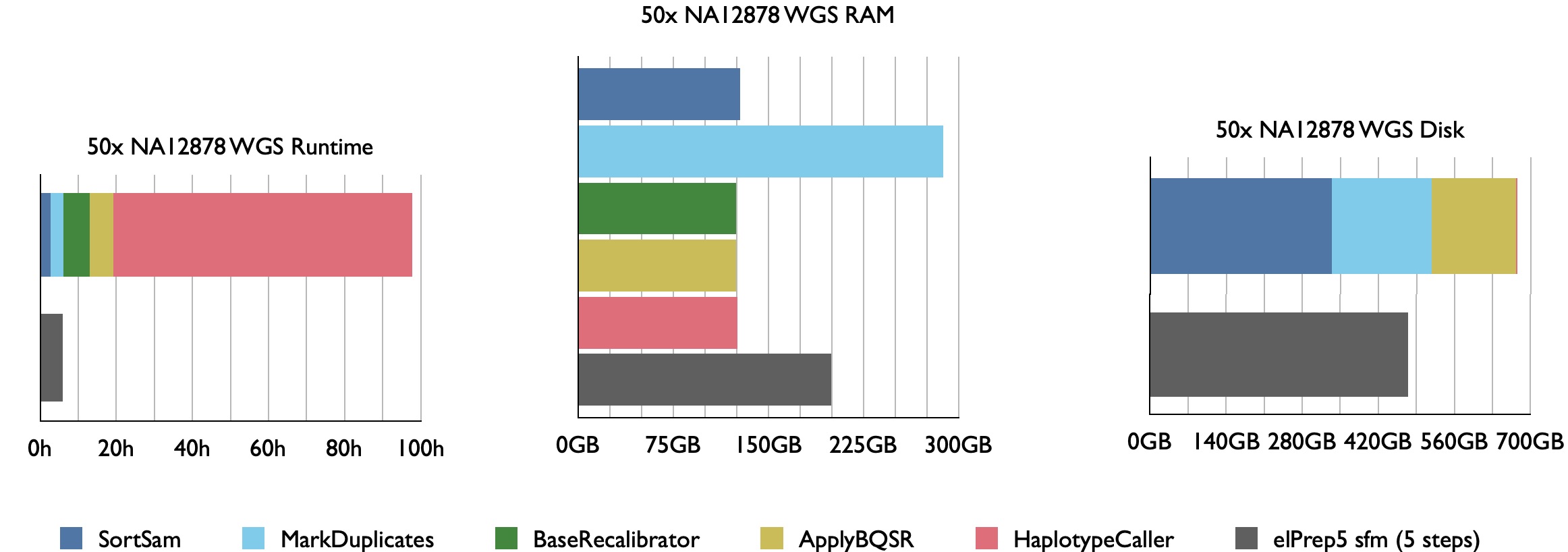
Our benchmarks show that elPrep 4.0.0 executes the sort/deduplicate/recalibrate and apply-BQSR-pipeline from the GATK Best Practices up to 12x faster for WES data and 7.5x faster for WGS data, while utilising similar or fewer compute resources than Picard/GATK4.
Example runtime, RAM use, and disk use for 50x WGS Illumina Platinum Genome NA12878 aligned against hg38. elPrep combines the execution of the 4 pipeline steps for efficient parallel execution.
We are looking forward to your feedback and suggestions.
Thanks a lot!
Kind regards,
Charlotte Herzeel, Exascience Life Lab, Imec, Belgium


Hi, this is a great tool! I feel you forgot to mention elPrep's modularity by design. Adding/removing filters to our elPrep call to suit our pipeline needs is done in a breeze. This allows us to use it in all sorts of NGS pipelines, and not just the GATK's Best Practices for variant calling. I also really like that -- with a bit of work -- it's not extremely difficult to add new filters to suit our needs. Plus, you guys have always been very responsive to such requests. This is a very efficient and valuable tool for the community. Thanks!
Thanks! elPrep is indeed designed as a modular plug-in architecture where the implementation of SAM/BAM tools is separated from the engine that parallelises and merges their execution. We have extensive documentation and very much welcome contributions and suggestions for extending elPrep to support different sequencing pipelines!
Thanks for the API documentation link!
Hi, I tried this some time ago, and found it made significant assumptions about read names. I.e. data from the SRA or non-illumina sequencers could not be processed. Have these requirements been relaxed in the meantime ?
Hi, We only make assumptions about the read names (QNAME) for optical duplicate marking, as they have to encode the tile + coordinates. Is this what you mean? If not, could you provide more details, e.g. the error message you get? Thanks!
If one does not
fastq-dumpdata from SRA with-F or --origfmtoption then one ends up with fastq headers that replace the standard Illumina headers with something that look like this.I believe that is what @colindaven is referring to. Then there are probably headers from other technologies that don't follow the Illumina format.
Yes, you are right. We have seen the same problem. elPrep currently only supports the Illumina format for optical duplicate marking, which is what GATK4 also supports by default. If you would like us to support other formats, please submit an issue on our github repository so we can discuss this in more detail. Thanks a lot.
(Edited: solved) looks like ePrep works in all other cases where there is no optical duplication
there is a lot of data in SRA where one cannot recover the original read formatting even if these were originally produced on that instrument.
Nonetheless, it's a nice tool and it's great people are trying to speed up bioinformatics infrastructure akin to what is going in the commercial and semi-commercial world with DRAGEN, MPEG-G and so on. So I will test a bit on Illumina X10 and NextSeq data I have and give feedback on any bugs I encounter.
Hi,
I am a bit confused about your remark. We tested elPrep a lot, including on data from SRA archives, but apart from optical duplicate marking, we haven’t encountered any issues because of QNAME fields. When elPrep is not able to recover tile information from the QNAME fields, it will skip optical duplicate marking and log a warning. Any other commands in the elPrep call should continue executing without problems.
There are two other places where elPrep code refers to QNAME fields. One is when sorting reads by queryname. The other is for correlating the two ends of a pair during duplicate marking, and for resolving ties when duplicates have the same phred score. To the best of our knowledge, we are in both cases faithfully reproducing the behaviour of Picard and GATK. We think that even for optical duplicate marking, if Picard sees QNAME fields without tile information, it will also not be able to properly mark optical duplicates.
We are primarily software engineers, so it is certainly possible we may be missing something. If you could clarify what the issue is you are referring to, we are very happy to make an attempt at fixing it.
Thanks a lot for your help.
I was simply reacting to your statement above where you say:
That is a much more restrictive statement than the second statement that you make in your reply:
If you support an Illumina specific functionality (among the many others) that does not mean that the tool "only supports Illumina format". Frankly, there is not even such a thing as "Illumina format", it just happens that for the past few years the most popular instruments produced read names formatted in a certain way, but that is not really a format, nor did Illumina instruments always produced that format.
If the tool works fine on say PacBio data, other than optical marking (which would not even apply there anyway) then it is all good and it is a fair replacement for samtools.
The original elPrep paper describes the sorting and duplicate marking implementations.
Is there a paper in preparation describing the BQSR implementation and the new features?