Hi,
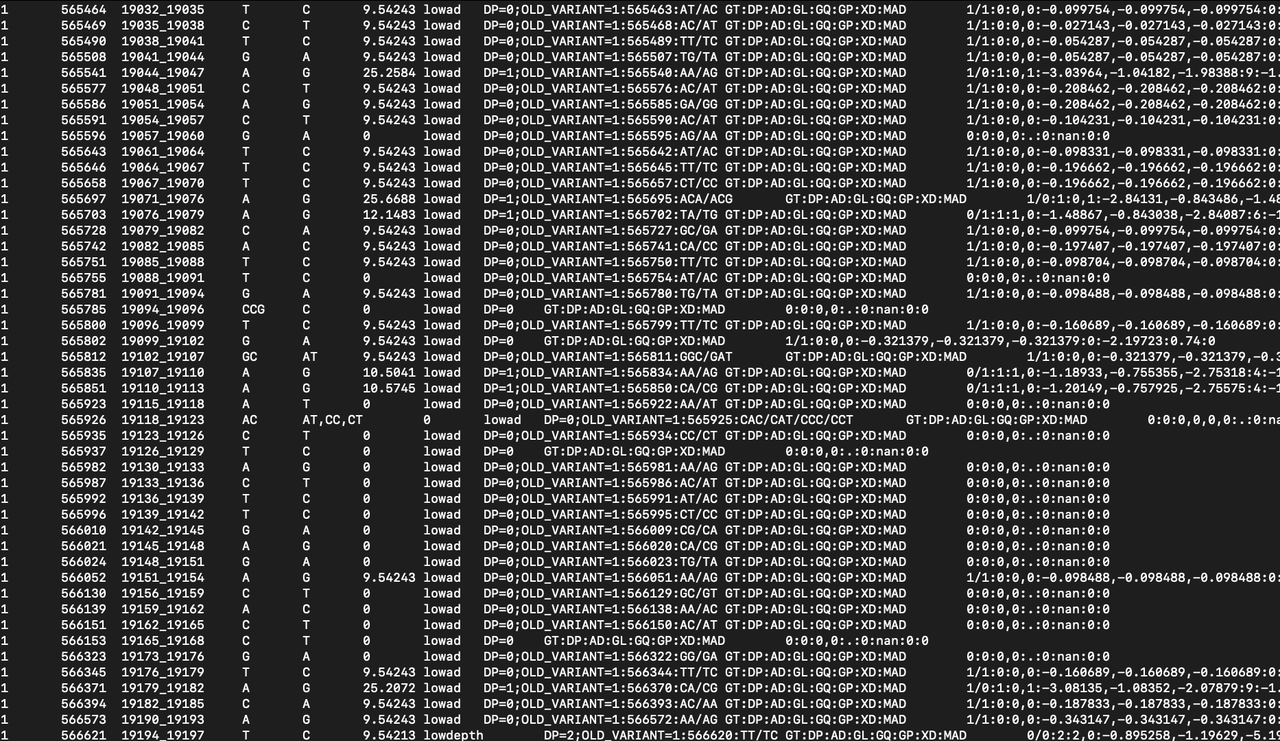
I am working with a VCF (See screenshot) created from a gam file. In my case I am using an option that would call every covered position but as you can see in position "565982" some of them don't have a genotype. I would like to know if there is an easy way to filter my VCF recovering only my position with a genotype called ? I was thinking about doing my own script that would parse the VCF but pretty sure something is already out there doing it, but I can't find it.
Thanks a lot
PS: without using this option I would have "normal" VCF but the call 0/0 are not reported and I want to recover those aswell in my case.


Thanks a lot for your quick tips. It worked as expected and way quicker than coding my own one ! Thank you !