Entering edit mode
3.7 years ago
javanokendo
▴
60
I have done the GSEA on my RNAseq data and I got the ridgeplot from the clusterProfiler bioconductor package. Could you guys help in interprating this figure?
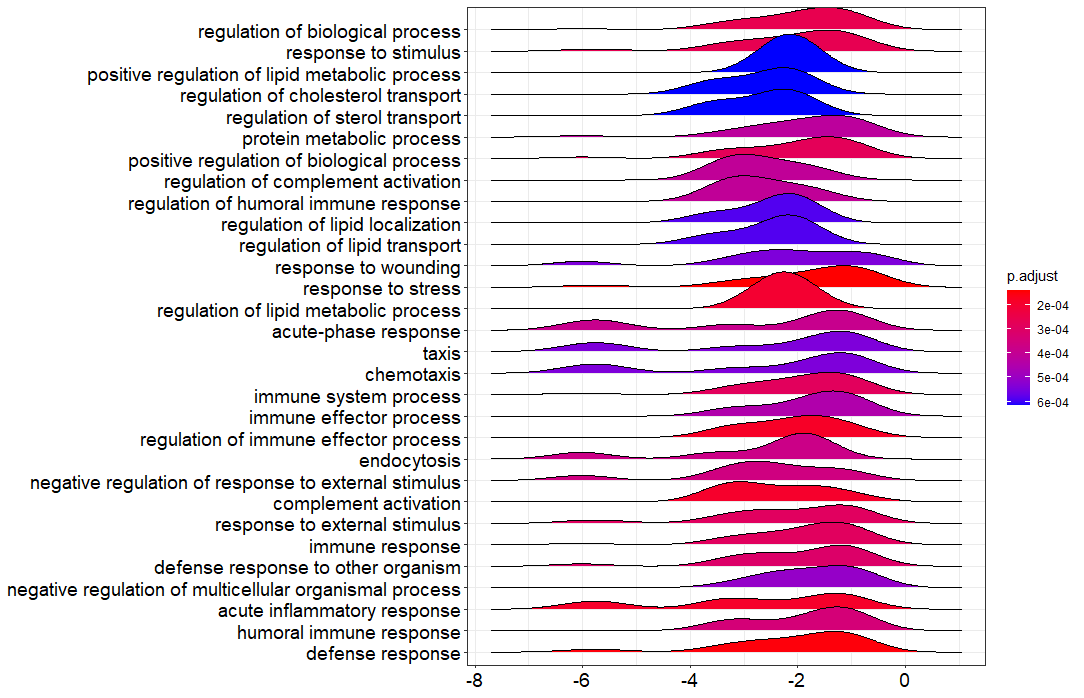


Thank you for your comment. These are the LFC values indicated on the X-axis. I have the following concerns and i hope you will help:
X-axis indeed indicates LFC values as reported earlier. The color describes the FDR/adjusted p-value as listed in the color bar, so only notifies you whether an enriched term is significant or not (e.g., p-value cutoff <0.05). To see whether a term is activated or suppressed you can use the enrichment score or NES (positive NES score == activated, negative NES == suppressed). The shape describes the density/number of genes with a specific LFC value, so it shows you the LFC distribution of genes in the gene set of an enriched term.
I now have a clear understanding of this result. Initially I thought that the X-axis values were just absolute numbers but I have realized that they have to be used in the interpretation of the results.