I used multiple structural variant(SV) callers to call SVs for paired end sequencing data.
Which genome browser offers the best support to visually inspect paired en sequencing data. I want to load the mapped paired end sequencing bams and then visually inspect the regions the SV callers marked as having structural variations.

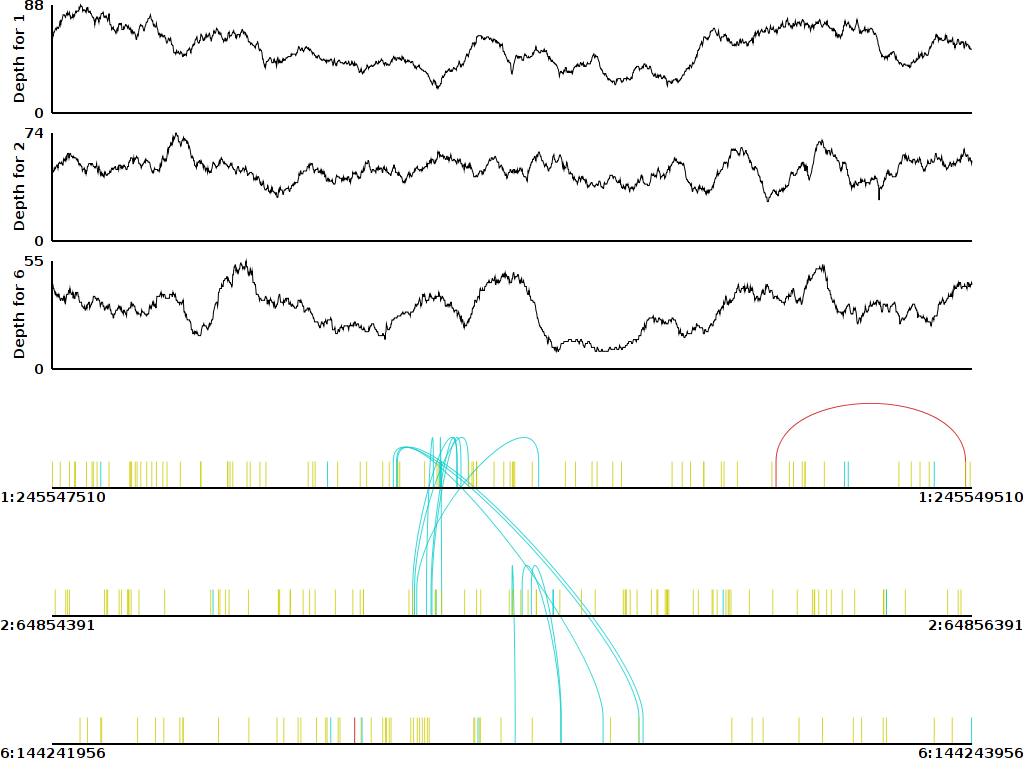


I was about to reply 'IGV', but I've found IGV very difficult to use when you want to visualize the SVs (especially when you have to compare several BAMs on the same screen)