I'm trying to figure out what Ensembl means by translated exons, when these are annotated for a gene based on Ensembl API. I notice that the translated exons associated with a transcript are sometimes different from the exons in the transcript. Why is that? Why aren't the translated exons of a transcript a strict subset of the exons in the transcript? Thanks.
Similar to JC's answer.
In some gene annotation schemes a distinction is made between: (1) exon coordinates of a transcript without regard to coding, and (2) exon coordinates for only the coding portion (i.e. the translated portion).
If the ORF starts within the boundaries of an exon (not right at the beginning), and you represent the coding/translated blocks independently of the exon blocks, you will have the first and last coding exon sharing one but not two coordinates with transcript exons. For all internal exons the coordinates will match up.
This concept, used in Ensembl is similar to that of the GTF file format. Perhaps examining the coordinates of a simple gene in GTF format will be helpful. Four types of coordinate blocks are defined: 'exon' (full exon), 'CDS' (coding portion of exon), 'start' (start codon), and 'stop' (stop codon). Note how for exons 1 and 6 the coordinates for 'exon' and 'CDS' match at only one end. For exons 2-5, the coordinates for 'exon' and 'CDS' match at both ends.
chr3 hg19_refGene exon 124449213 124449474 0 + . exon "1"; gene_id "NM_000373";
chr3 hg19_refGene CDS 124449319 124449474 0 + 0 exon "1"; gene_id "NM_000373";
chr3 hg19_refGene start 124449319 124449321 0 + . exon "1"; gene_id "NM_000373";
chr3 hg19_refGene CDS 124453940 124454093 0 + 0 exon "2"; gene_id "NM_000373";
chr3 hg19_refGene exon 124453940 124454093 0 + . exon "2"; gene_id "NM_000373";
chr3 hg19_refGene CDS 124456415 124457086 0 + 2 exon "3"; gene_id "NM_000373";
chr3 hg19_refGene exon 124456415 124457086 0 + . exon "3"; gene_id "NM_000373";
chr3 hg19_refGene CDS 124458871 124459046 0 + 2 exon "4"; gene_id "NM_000373";
chr3 hg19_refGene exon 124458871 124459046 0 + . exon "4"; gene_id "NM_000373";
chr3 hg19_refGene CDS 124460999 124461113 0 + 0 exon "5"; gene_id "NM_000373";
chr3 hg19_refGene exon 124460999 124461113 0 + . exon "5"; gene_id "NM_000373";
chr3 hg19_refGene CDS 124462762 124462928 0 + 2 exon "6"; gene_id "NM_000373";
chr3 hg19_refGene stop 124462929 124462931 0 + . exon "6"; gene_id "NM_000373";
chr3 hg19_refGene exon 124462762 124468119 0 + . exon "6"; gene_id "NM_000373";
5' and 3' UTRs are considered exonic because they are not explicitly spliced out.
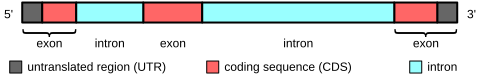
Because biology says that. When a gene is transcribed, a typical mRNA includes the coding exons and other segments as signals and regulatory elements. Those elements are not transcribed into protein, but are exonic elements too.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Can you provide a specific example - a pair of links in Ensembl?