Hi, everyone, I work on microbial genomes.Do anyone know an all in one software which can be useful for me to study all aspects of bacterial genome data?
A single software which is able to perform various tasks like multiple genome alignments, phylogenetic studies , genome visualizations, SNP analysis, *core and dispensable genome visualizations and many such things* will be highly acceptable.
I can manage funds upto 2500 USD.
Thanks in advance.
I think finding one program to do it all is the easy way out: You'll understand more and have more control if you use specific tools designed for each respective job.
That being said, if I was to recommend a single program I think the CLC bio platform (genomics workbench or main workbench) is quite good. The genome alignment and visualization aspects might be a bit lacking for you compared to other resources, but it's basically a complete package. There are plugins to add to its functionality.
You might be interested in a comparable package called Lasergene from DNAstar.
However I agree with Josh that individual tools might be better to tune what you actually want.
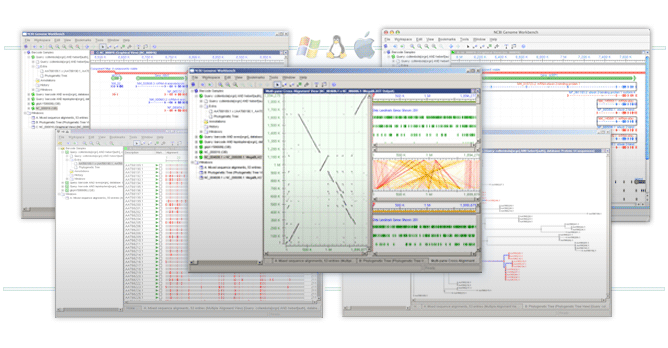
Have you checked out the free NCBI Genomics Workbench and Ugene already? They can do most as well.
what I meant by 'core and dispensable genome visualizations' is that, Creating Ortholog clusters from many genomes same as CD-HIT server also plot fall in number of core genes and rise in dispensable genes as each new genome is added to the dataset. Same as this image for example: http://genomebiology.com/2007/8/5/R71/figure/F2
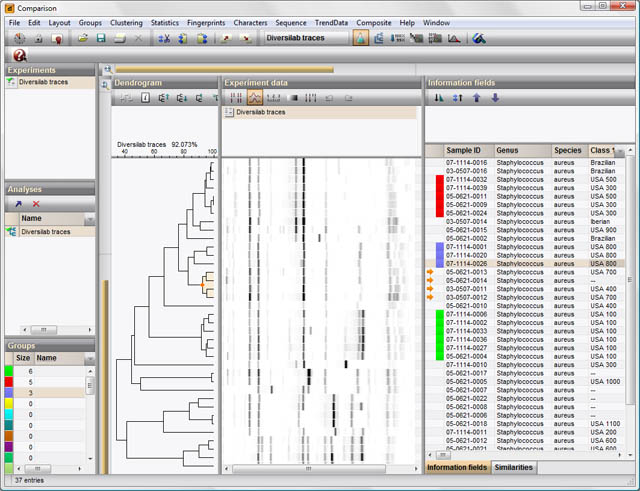
BioNumerics is the all-in -one solution you are looking for (disclaimer: I work for Applied Maths, the company that develops BioNumerics). It is extensively used for studying all aspects of bacterial genome data. Specifically, for the areas you are interested in:
-
multiple genome alignments: "The Genome Analysis Tools module offers side-by-side comparison of genomes and chromosomes, analysis of organization and functional behavior of genomes, alignment of multiple chromosomes and chromosome-based SNP analysis."
-
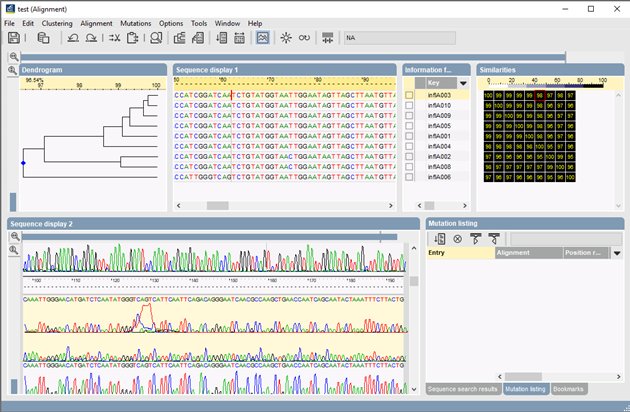
phylogenetic studies: "BioNumerics offers Maximum Parsimony and Maximum Likelihood as phylogenetic inference methods. Besides standard algorithms, the optimal trees can be calculated using simulated annealing or quartet puzzling. Both methods result in an unrooted tree, which can be converted into a rooted tree after assignment of a root. To correct phylogenetic distance scaling, the Jukes & Cantor or Kimura 2 parameter correction factors can be chosen."
-
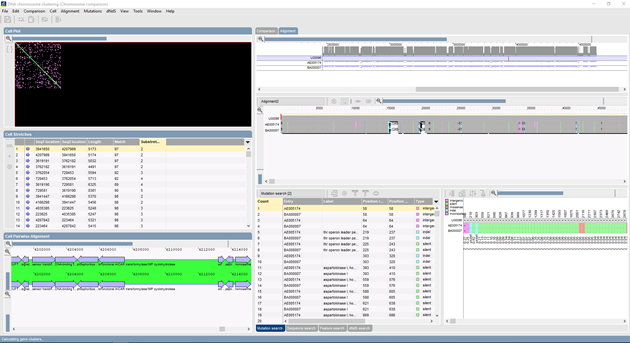
SNP analysis: "BioNumerics’ sequence alignment application is an invaluable tool for SNP and mutation analysis. SNPs or mutations are screened for through up to many thousands of aligned sequences. The software statistically calculates the probability of each SNP based upon the quality of the base assignments and the curves in the chromatogram files. Various filters allow for screening SNPs with specific thresholds or other features such as the type of mutation they induce. By looking at sequencer chromatograms directly, the user has excellent control over the probability of each potential SNP locus. Complete alignments, including all SNP and subsequence search listings, can be saved and re-edited at any time. Additional sequences can be added to existing projects."
I am not completely sure what you mean by 'core and dispensable genome visualizations', but if there is something you really need/want that currently is not available in the software, then we can always write a custom script for you to add that functionality.
Please contact our sales department for a free evaluation software license or to request a quote.
That looks pretty nifty! The truth is that when it comes to visualization there are a lot fewer open source options - and the commercial solutions always seem better.
I think it has to do with the fact that open source thrives when the quality metrics are "objective": Faster, more accurate, fewer resources.
When it comes to what looks good or makes sense the metrics are subjective and it is not immediately obvious which option is better - hence the open source meritocracy does not work that well.
@Jeroen: Of course I need all above functionality, (in addition) what I meant by 'core and dispensable genome visualizations' is that, Creating Ortholog clusters from many genomes same as CD-HIT server also plot fall in number of core genes and rise in dispensable genes as each new genome is added to the dataset. Same as this image for example: http://genomebiology.com/2007/8/5/R71/figure/F2
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


This very much depends on your bioinformatic skills - and backing.
Galaxy ( https://main.g2.bx.psu.edu/ ) ? But it's free...