Hi everyone,
I post today to ask for help in order to use ggplot2 package, but first the fundamental question:
Is possible to join in one plot multiple trendlines of different datasets?
The reason of this question is because I need to visualize in the same plot, the ratio of reads between 3 chipseq samples and 1 input using a trendline for each sample in a big intronic region. So far I've tried to do this with GenomeGraph R package but it was confusing for me to get this plot.
So I decided to do it with ggplot2 package. The input is a file that have almost 300 bins of this big intron and for each bin the number of reads of each sample:
iD pol.t0 pol.t5 pol.t60 input
Intron279 11 10 5 17
Intron278 10 24 20 30
Intron277 17 58 20 41
Intron276 5 29 9 38
Intron275 5 16 6 29
Intron274 4 14 10 28
Intron273 11 33 17 28
Intron272 11 40 15 36
Intron271 8 18 4 36
Intron270 7 24 5 32
And this is my Rscript to plot the final trends in a separate plot each:
library(ggplot2)
t<-read.table("BigIntron_reordered.bed")
## Ratio Calculation
t2<-data.frame(t[,1], (t[,2]/t[,5]), (t[,3]/t[,5]), (t[,4]/t[,5]))
colnames(t2)<-c("iD","pol.t0","pol.t5","pol.t60")
## No lose initial order of iDs
t2$iD<-factor(t2$iD, levels=unique(as.character(t2$iD)) )
## Plot trendlines for each sample
p1<-ggplot(t2[t2$pol.t0 != "Inf" & t2$pol.t0 != "NaN",], aes(x=iD, y=pol.t0)) + stat_smooth(method="loess",aes(group=1),size=2)
p2<-ggplot(t2[t2$pol.t0 != "Inf" & t2$pol.t0 != "NaN",], aes(x=iD, y=pol.t5)) + stat_smooth(method="loess",aes(group=1),size=2)
p3<-ggplot(t2[t2$pol.t0 != "Inf" & t2$pol.t0 != "NaN",], aes(x=iD, y=pol.t60)) + stat_smooth(method="loess",aes(group=1),size=2)
## Grid plot of everything together
grid.arrange(p1, p2, p3, nrow=3)
The final result is:
So is possible to join this trend lines all together in one plot?
Thanks for your help!!


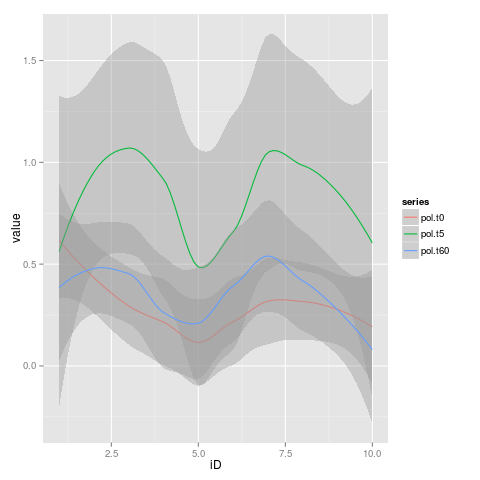

Yes, sure it is for that! Thanks for your answer was perfectly clear!