Dear all,
We have got RNA-Seq of a plant without a reference genome, so we de novo assembled its transcriptome using Trinity. Now we have an annotation file of this transcriptome. I want to generate GO (Gene Ontology) functional classification using Blast2GO. But re-mapping my Blast XML results in Blast2GO is time consuming (I tried that of my ~200,000 transcripts in my transcriptome).
An alternative way to generate GO function classification is importing the annotation file (with a suffix .annot according to Blast2GO manual), the .annot file looks like below (You can also get this example .annot file at http://www.blast2go.com/b2glaunch/resources, named b2g_example_files.zip): p.s. If you can't see this picture from Flickr, I also put it on GitHub
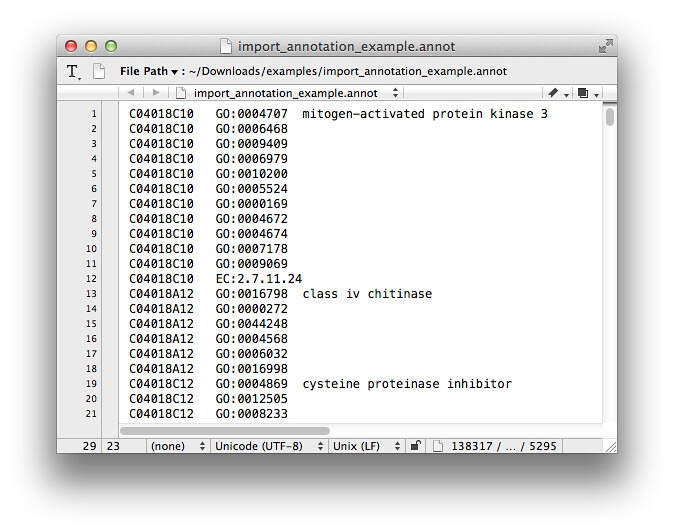
But our annotation file was not generated by Blast2GO, it's a CSV file like this below (a little mess):
X01_query_id, X06_hit_title, X07_molecular_function, X08_biological_process, X09_cellular_component ##header of the CSV file
comp1000113_c0_seq1, Cc-nbs resistance protein [Medicago truncatula], GO:0043531 // ADP binding;GO:0005524 // ATP binding;GO:0017111 // nucleoside-triphosphatase activity, GO:0006952 // defense response,
comp10001_c0_seq1, Pistil-specific extensin-like protein [Medicago truncatula], , ,
comp1000255_c0_seq1, F-box protein [Medicago truncatula], , ,
comp1000736_c0_seq1, Alpha-L-arabinofuranosidase [Medicago truncatula],GO:0046556 // alpha-N-arabinofuranosidase activity, GO:0046373 // L-arabinose metabolic process,
comp1000860_c0_seq1, Protein kinase [Medicago truncatula], GO:0005524 // ATP binding;GO:0004674 // protein serine/threonine kinase activity, ,
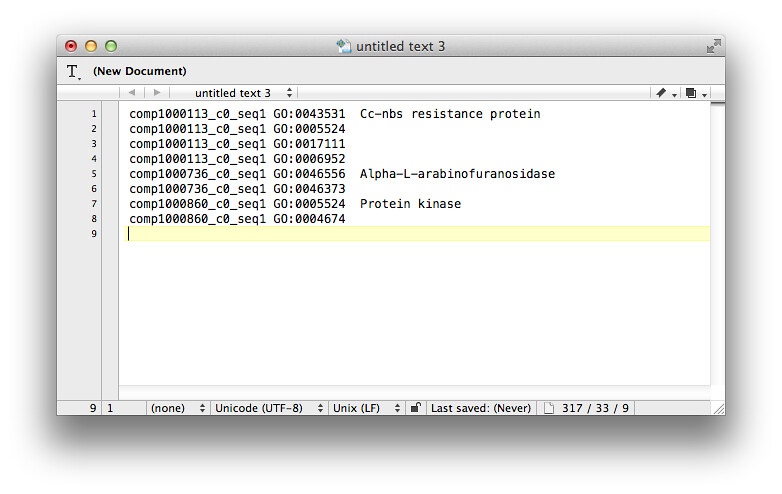
I need to bother you to help me provide some command or some scripts to generated a .annot file like this below:
Ignore those lines without GO IDs
p.s. If you can't see this picture from Flickr, I also put it on GitHub
I look forward to hearing from all of you soon.
Thank you and best regards,
lzsph


Hi Whetting,
Awesome. By the way, how to keep one protein name in each set? Like this below.
comp1000113c0seq1 GO:0043531 Cc-nbs resistance protein
comp1000113c0seq1 GO:0005524
comp1000113c0seq1 GO:0017111
comp1000113c0seq1 GO:0006952
Thank you very much!
Regards,
Lzsph
I edited the code...this should give you what you want
Hi Whetting, thanks again. It's perfect!!!