Hello everybody. Making a poster, I had the need to represent graphically a series of genes with their splicing in a simple way. I have a series of cDNA variants aligned against their correspondent genomic strand, and I was thinking about summarising the information displaying each genomic region as a thin line, the exons as thicker parts on the genomic region and the introns as graphical hairpins connecting exons (nothing new, very classical way). Is there a software that, given an alignment between a genomic region with its cDNAs (or at least some of them), outputs something like this? I started to do it manually, but the process is both slow and inaccurate.
Software To Graphically Represent The Splicing In A Simple Way
1
Entering edit mode
11.5 years ago
Anima Mundi
★
2.9k
2
Entering edit mode
11.5 years ago
Pierre Lindenbaum
164k
See also
Batch Viewing of UCSC Browser Graphic
here is a XML returned by the UCSC for the table knownGene:
$ mysql --user=genome --host=genome-mysql.cse.ucsc.edu -A -D hg19 -X -e 'select * from knownGene limit 2'
<resultset statement="select * from knownGene limit 2
" xmlns:xsi="<a href=" http:="" www.w3.org="" 2001="" XMLSchema-instance"="" rel="nofollow">http://www.w3.org/2001/XMLSchema-instance">
<row>
<field name="name">uc001aaa.3</field>
<field name="chrom">chr1</field>
<field name="strand">+</field>
<field name="txStart">11873</field>
<field name="txEnd">14409</field>
<field name="cdsStart">11873</field>
<field name="cdsEnd">11873</field>
<field name="exonCount">3</field>
<field name="exonStarts">11873,12612,13220,</field>
<field name="exonEnds">12227,12721,14409,</field>
<field name="proteinID"></field>
<field name="alignID">uc001aaa.3</field>
</row>
<row>
<field name="name">uc010nxr.1</field>
<field name="chrom">chr1</field>
<field name="strand">+</field>
<field name="txStart">11873</field>
<field name="txEnd">14409</field>
<field name="cdsStart">11873</field>
<field name="cdsEnd">11873</field>
<field name="exonCount">3</field>
<field name="exonStarts">11873,12645,13220,</field>
<field name="exonEnds">12227,12697,14409,</field>
<field name="proteinID"></field>
<field name="alignID">uc010nxr.1</field>
</row>
</resultset>
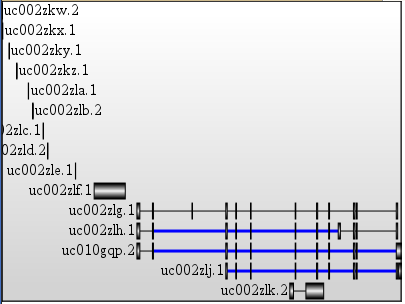
if you write your data in that format, you can process the XML file with my XSLT stylesheet to produce a SVG file
$ mysql --user=genome --host=genome-mysql.cse.ucsc.edu -A -D hg19 -X -e 'select * from knownGene limit 2' | xsltproc <(curl -s "https://raw.github.com/lindenb/xslt-sandbox/master/stylesheets/bio/ucsc/ucsc-sql2svg.xsl") - | head
<svg:svg xmlns:svg="<a href=" http:="" www.w3.org="" 2000="" svg"="" rel="nofollow">http://www.w3.org/2000/svg" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xmlns:xlink="http://www.w3.org/1999/xlink" width="900" height="41" style="stroke-width:1px;">
<svg:title>select * from knownGene limit 2 </svg:title>
<svg:defs>
<svg:linearGradient x1="0%" y1="0%" x2="0%" y2="100%" id="grad">
<svg:stop offset="5%" stop-color="black"/>
<svg:stop offset="50%" stop-color="whitesmoke"/>
<svg:stop offset="95%" stop-color="black"/>
</svg:linearGradient>
<svg:linearGradient x1="0%" y1="0%" x2="0%" y2="100%" id="vertical_body_gradient">
and a result visualized in firefox:
2
Entering edit mode
11.5 years ago
Charles Warden
8.3k
I have heard that the sushimi plot is a new, useful way to visualize splicing events:
http://genes.mit.edu/burgelab/miso/docs/sashimi.html
I haven't had a chance to try out the software yet - let me know what you think!
Similar Posts
Loading Similar Posts
Traffic: 1930 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


this has already been asked previously. See How to visualize splice junction data, in particular exon-skipping, Data visualisation -- exon/intron map?