I'm looking for a tool (or suggested code) on how to illustrate a multiple sequence alignment in a very coarse manner, so that it can fit in a small figure. The point isn't to show specific amino acids, just to show where there are large gaps in the alignment. Really it's inspired by this image:

Any suggestions or pointers to code (eg matplotlib) would be greatly helpful. Thanks!

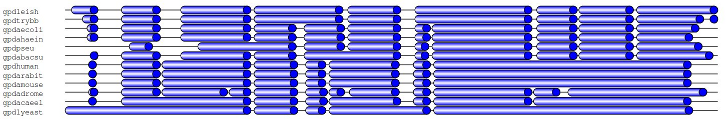

and Tim Yates wrote a groovy version: https://twitter.com/tim_yates/status/359990843546935297