Entering edit mode
11.2 years ago
Cacau
▴
520
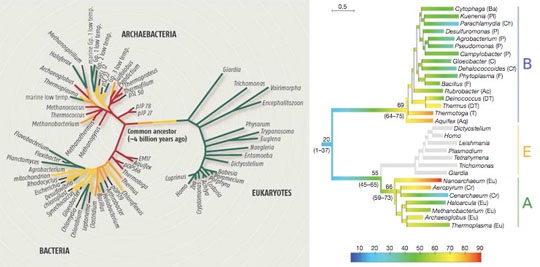
Does anyone have any idea about how the following figure (especially the right one) is drawn? I think the gradual change of the color within one branch mighe be a difficulty in drawing the figure.


if your picture is already available as a vectorial drawing, open it in inkscape an use the gradient tool: http://inkscape.org/doc/basic/tutorial-basic.html