Hi,
I was wondering if anyone knows of an easy web browser application/code that I could use to show small contigs/alignments (few Kb) and ideally allow editing, modifying the original file (or a copy).
Thanks.
Hi,
I was wondering if anyone knows of an easy web browser application/code that I could use to show small contigs/alignments (few Kb) and ideally allow editing, modifying the original file (or a copy).
Thanks.
"modifying the original" you meant the image ?
You could just encode your contigs as XML and transform it with XSLT to produce a SVG file. See that example with the UCSC data (2006) : http://plindenbaum.blogspot.fr/2006/02/ucsc-genome-browser-svg.html

and that previous post:
Batch Viewing of UCSC Browser Graphic
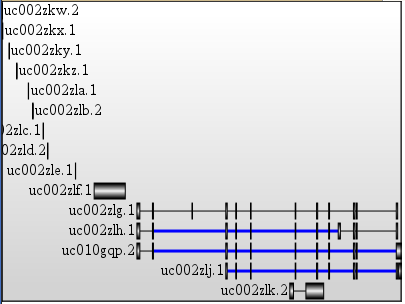
and that one : a Local file that can be edited; http://plindenbaum.blogspot.fr/2010/08/tiny-genome-browser-xhtmljavascriptjson.html
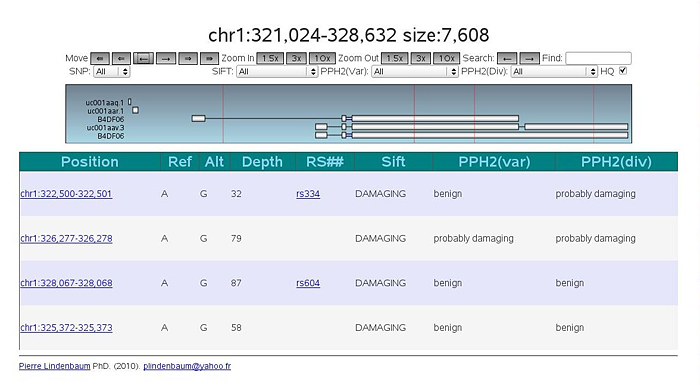
If you are familiar with Gbrowse, you can use Jbrowse and you can embed in your webpage.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.
By modifying the original I mean editing the alignment and changing the original file (fasta, phyllip...) if the user deletes an indel or moves a nucleotide.