Entering edit mode
3.5 years ago
xiaoleiusc
▴
140
Hi, All,
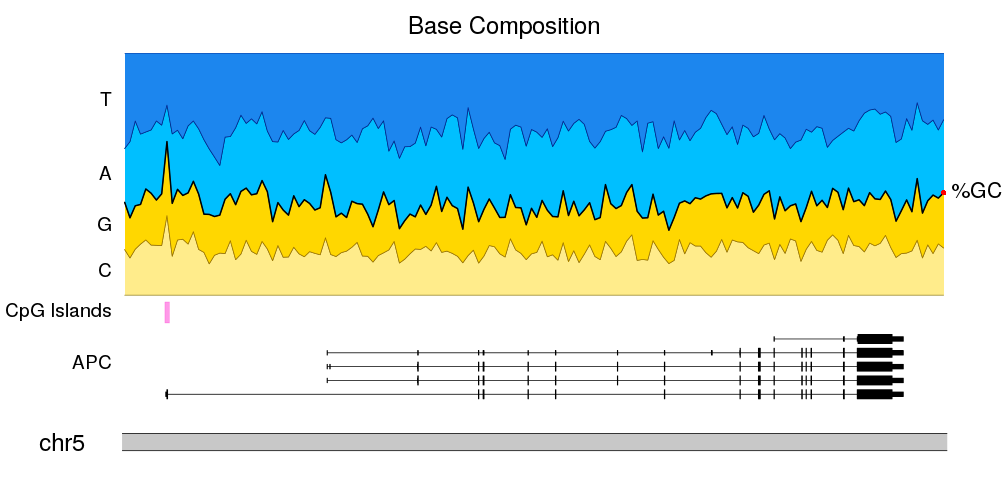
I wonder if there is any tool to do nucleotide composition analysis to generate figures such as this (link here)? It would be great if the sliding window size could be customized.
Thanks,
Xiao


Visualize nucleotides for every position in R