Entering edit mode
10.8 years ago
Scot Federman
▴
20
Hi - does anyone have a reference for the NCBI nt database size over time? It's surprisingly difficult to find this information on NCBI (or anywhere else online).

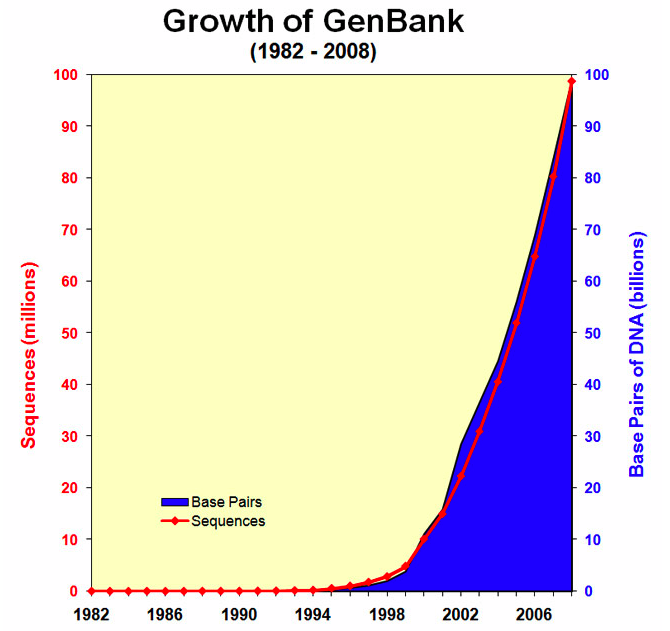

Thank you, Gergana - this is also all we could find online. Parsing the historical release notes (ref) is a good way to complete this plot.
However, apparently - nt is comprised of (Genbank + EMBL + DDBJ + PDB + RefSeq). I wonder if the only way to show nt size over time is to gather data from all of these sources?
I would assume that most of the data from the different databases will be overlapping and the most rich database will be that from Genbank. But even if you could collect non-reduntant dataset combining all 4 databases, that wouldn't change the overall trend. Unless you would like to be super precise about the numbers...
Since the International Nucleotide Sequence Database Collaboration (INSDC), which comprises DDBJ, ENA and GenBank, ensures that the major nucleotide sequence databases contain the same entry data, and RefSeq is derived from the contents of GenBank the volume of data in 'nt' is directly related to the size of the INSDC databases.
That you can reasonably assume that the size of 'nt' approximates that of the INSDC member databases. So see their statistics pages:
The database sizes are commonly reported in the associated release notes for each database as well.
If you require exact figures for the size of NCBI's 'nt' database, I suggest you ask the NCBI help-desk (see http://www.ncbi.nlm.nih.gov/About/glance/contact_info.html) for figures from their records.