Entering edit mode
3.5 years ago
ashwing.kofficial
▴
10
Hello guys,
I just finished my undergrad project which consists of pathway study of a disease using Rna-seq data. To publish the paper, I though I could use a few images for the analysis. Is there any good visualization software for Rna-seq data where I can use my degs for visualization.


I know of at least two tools to visualize enriched pathways: DAVID , which is a bit old but still useful last time I checked, and reactome.org which provide really nice illustration of protein pathways.
I have used DAVID before but it is not capable of visualization tools, but gives a detailed analytical report. I am looking for visuals which represent an overall aspect. Also,Reactome is more specific than DAVID
You can visualize pathway enrichment with DAVID. It shows DE genes (highlighted with a red star) on full KEGG pathways, for instance like this: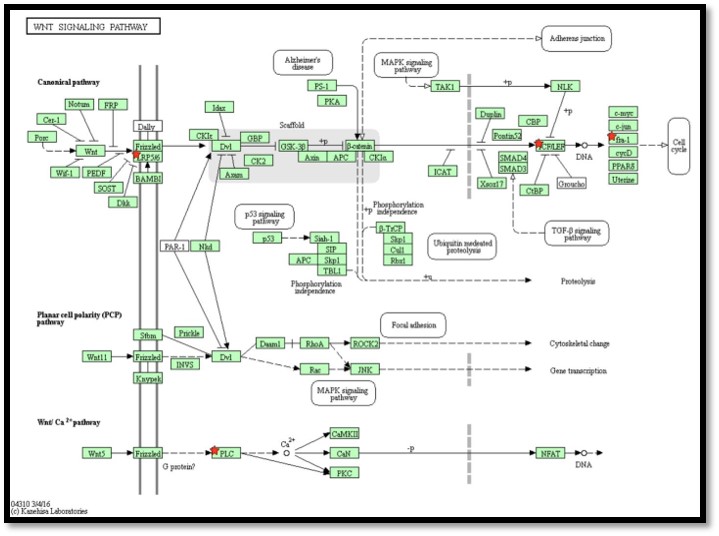
Is it what you are looking for or were you thinking of a different kind of "images for pathways analysis".
No sir, I already have that, I am looking for an image which can illustrate the DEGS in a summarized manner.
Sorry I misunderstood, but then it is still unclear for me what you are looking for.