Entering edit mode
4.3 years ago
DN99
▴
20
I'm plotting an upset plot of KEGG enriched pathways using DOSE and clusterprofiler which works fine like this:
kk <- enrichKEGG(gene = gene_ids,
organism = 'hsa',
pvalueCutoff = 0.05,
pAdjustMethod = 'fdr')
upsetplot(kk)
The plot given looks like this:
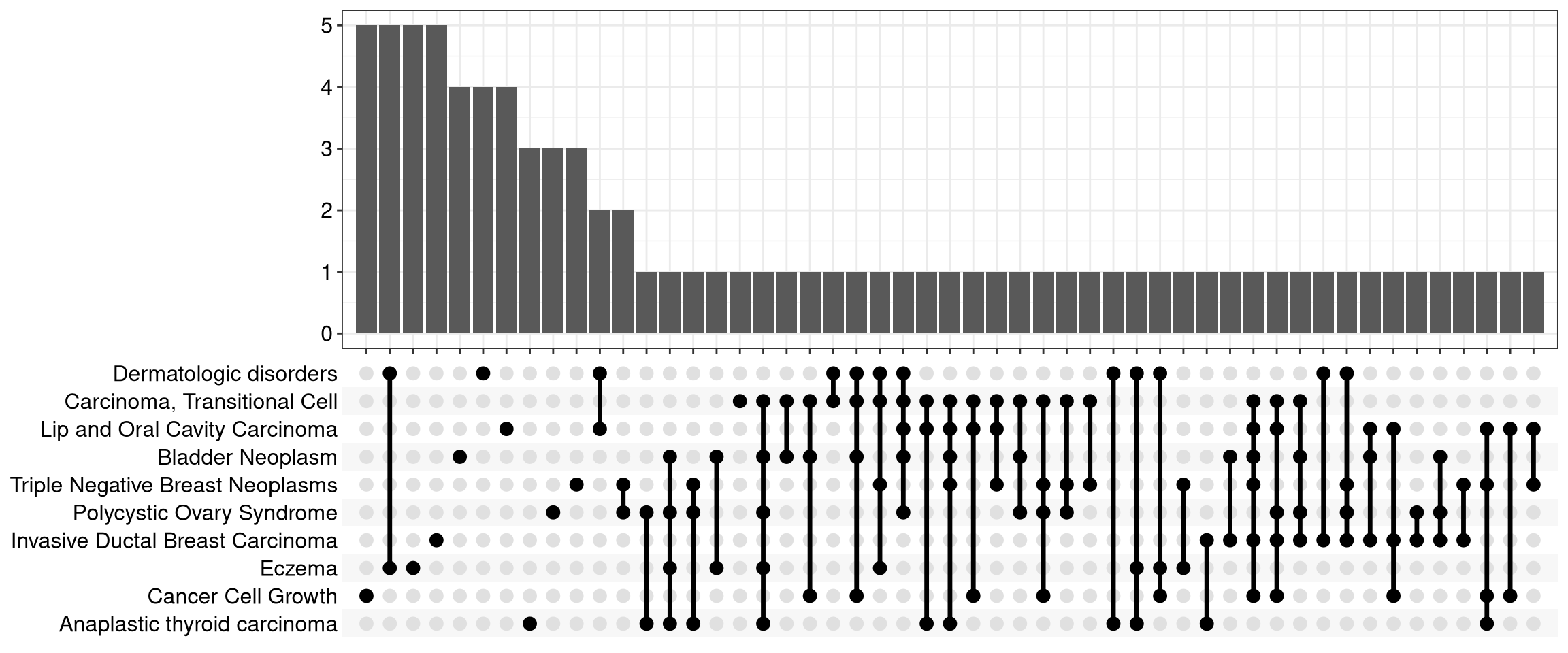
I'm wondering if there's a way to expand this plot on the left to have a bar chart of the number of genes enriched per pathway? So the style of the plot would expand to look like this:
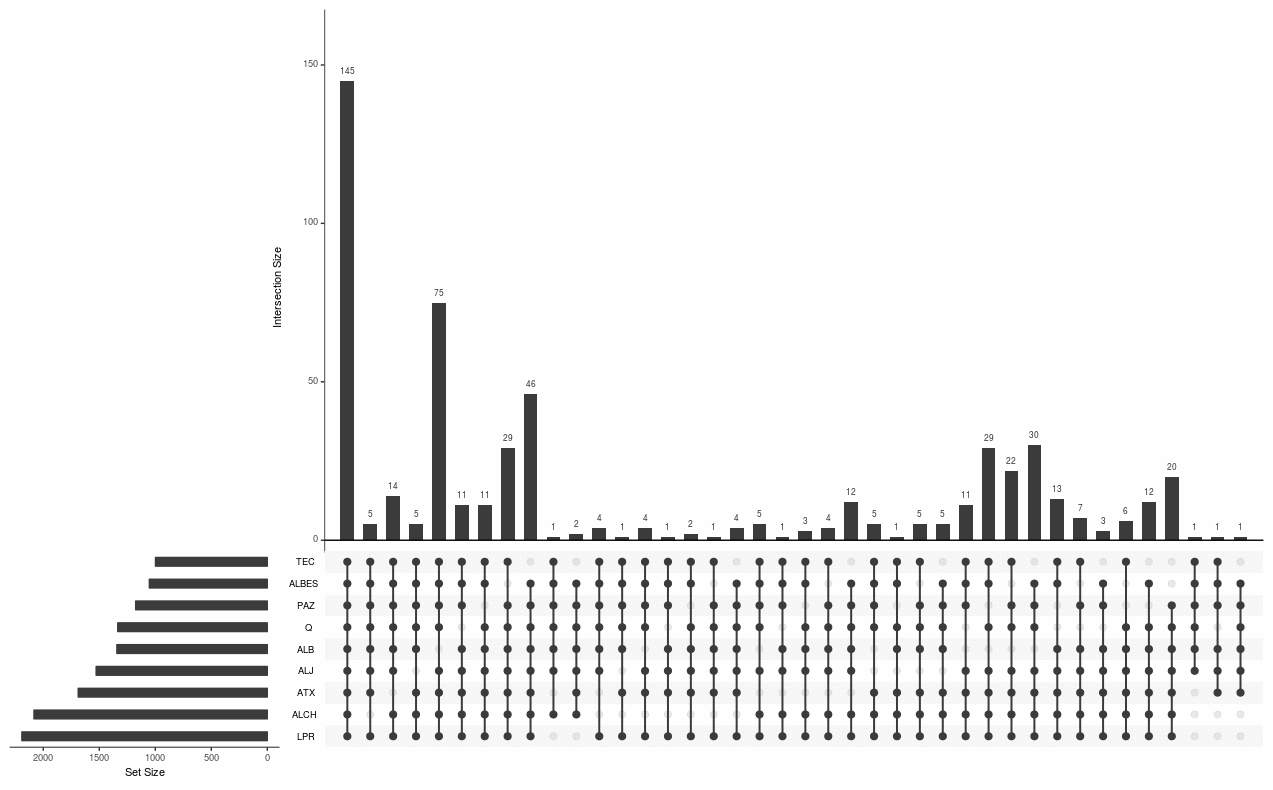
Is this possible using the upsetplot() function? Or would I need to try plotting the kk data with another package like UpSetR?

