Hei everyone,
for my project i did capture sequencing of mulitple individuums. Somehow the depth and coverage is not as high as expected. Now i want to plot the output of samtools depth. Unfortunatly i am not able to write a perfect code, so I thought i ask here for help. I would like to create via R on the server a histogramm. On the x-axis should be plotted the position in basepairs and on the y-axes the depth. The bins of the x-axis should conclude 1Mbases and at last it would be perfect if each bin shows its mean depth. I allready used samtools coverage but i would like to see also the visualization of samtools depth. I know it is a big request, but if you can help me i would be very greatful;). May you can also share here your codes you are using for your visualization.
Thanks a lot
Dominik

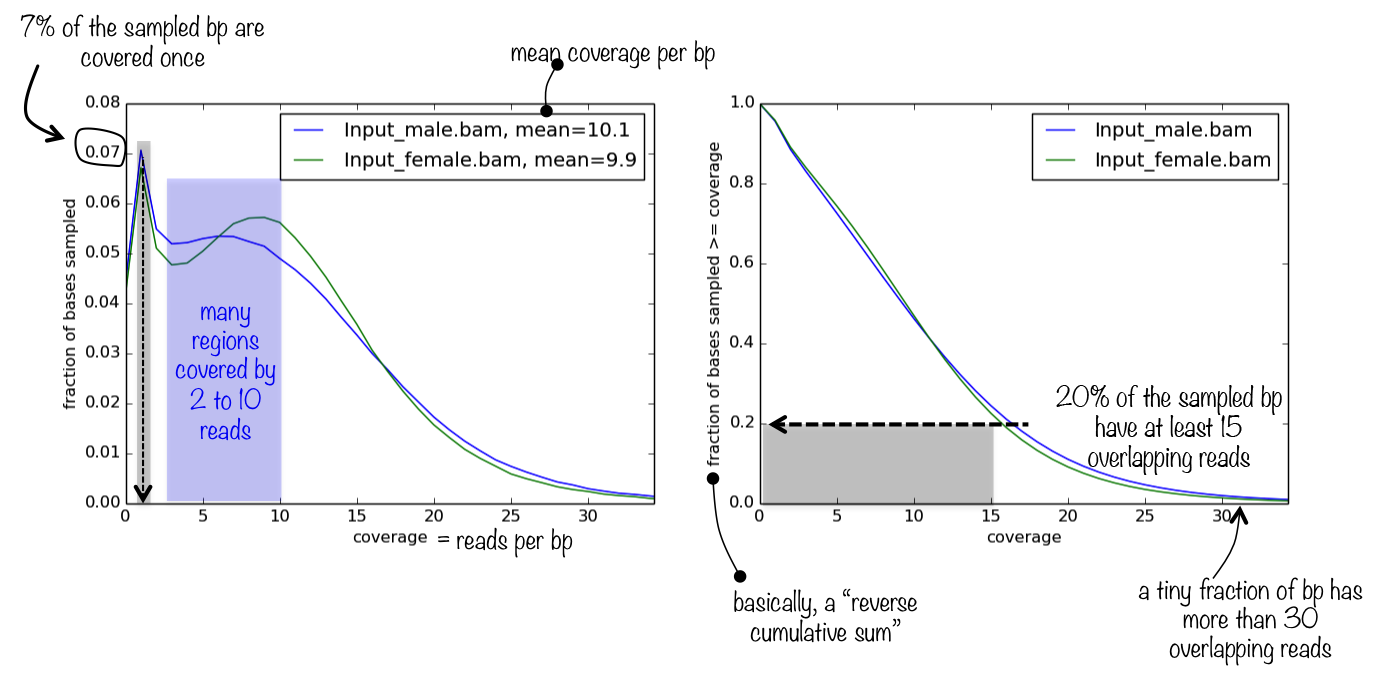


If you're using
samtools depthand the coverage isn't what you expect, it's likely the inbuilt limit of maximum depth (-m). This is because it uses the slow pileup engine. I completely rewrote the depth command for samtools 1.13 and so it's much much faster on very deep data and also no longer has the paradox of needing a maximum depth limit!Besides, upgrading to the latest version brings other benefits anyway.
As for plotting, personally I tend to use hacks of awk and gnuplot, but I'm a bit of a luddite like that. :-)