You'll have to bin the data to plot them, as has been pointed out in another comment here. Then you could calculate the frequencies of the -1s and 1s, and visualize that as a line plot. Here's an example implementation in R (the only external package used here is ggplot2 for the visualization). The code should be (hopefully) self-explanatory.
#Toy data.
x <- sample(c(1:9999999), 100000, replace = FALSE)
y <- sample(c(-1, 1), 100000, replace = TRUE, prob = runif(2))
df <- data.frame(x = x, y = y)
df <- df[order(df$x),]
#Script really starts here.
#Change bin_size to control, well, the bin size.
bin_size <- 100000
#Counters.
curpos <- 1
max_val <- max(df$x)
#Aggergating counts over chromosome positions falling within
#bins defined by curpos and endpos.
while(curpos <= max_val){
endpos <- curpos + bin_size
if(curpos == 1){
endpos <- endpos - 1
}
#Get all genomic coordinates in this window into a temporary data.frame.
curdf <- df[df$x >= curpos & df$x < endpos, ]
#And their counts also.
curdf <- as.data.frame(table(curdf$y))
curdf$startpos <- curpos
curdf$endpos <- endpos
#Calculating midpoint value of the bin (for plotting)
curdf$mid <- (curdf$endpos-curdf$startpos)/2 + curdf$startpos
#Collecting this output for every bin into outdf.
if(curpos == 1){
outdf <- curdf
} else{
outdf <- do.call("rbind", list(outdf, curdf))
}
cat("Working on bin ", curpos, " -- ", endpos, "\n")
#Updating position for next iteration.
curpos <- endpos
}
#Plotting
library(ggplot2)
ggplot(data = outdf, aes(x = mid, y = Freq, color = Var1)) +
geom_line(alpha = 1.0) + facet_wrap(~Var1)
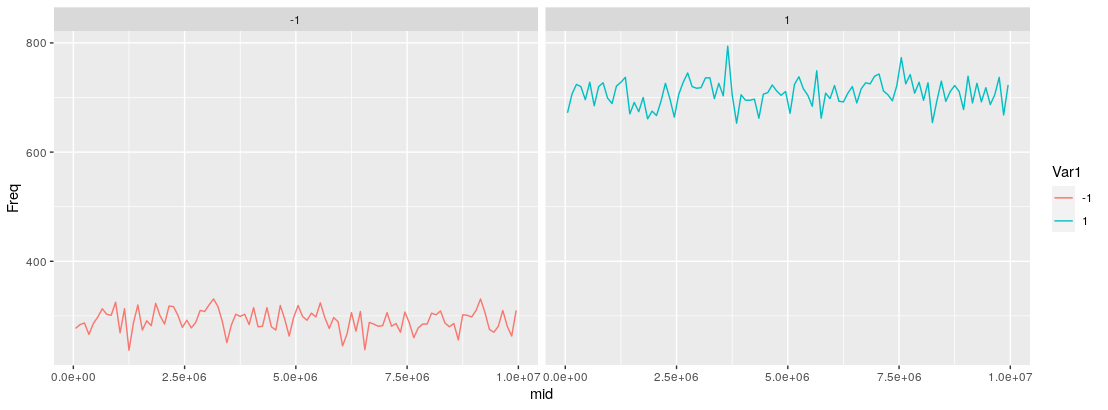
You could opt not to facet the output as has been done here, and just have the lines plotted in a single pane (just remove + facet_wrap(~Var1), and adjust the alpha in geom_line() if the lines overlap).
Was this what you were going for?



I have a script here that I think may handle that or could be easily modified to do it. It handles individual chromosomes. There's an example of three in an image; however, it can do one as well.
Minor point: You data example looks like maybe TSV and not CSV?
(Oh yes, meant to add that definitely if you have that many points, you'll have issues trying to plot them all. Do you need it all though? With the human genome, I think I sample in the demo notebook for my script 10% of the points and plot them to get something that doesn't exceed the limited memory of MyBinder.org session instances.)
which tool ? R ?