my sequences have names of more than 10 characters. but looks like phylip format takes only names with characters less than 10. How to deal with it ??? I tried shortening the names but cannot do that for all sequences
I assume you have the sequences in a FASTA file. I recommend to shorten the sequences in FASTA format already, then convert to phylip format using EMBOSS tool seqret.
I think there should be many scripts written by folks doing phylogenetics to cope with header length issues. Here is one I just found https://github.com/nylander/translate_fasta_headers It seems it can do what Mensur suggests and then rebuild the original sequence ids in the Newick output.
Phylip format can have an arbitrary number of characters in header, but not all the programs will tolerate it. MrBayes, for example, has no complaints when headers are longer.
but cannot do that for all sequences
Of course you can. Each name can be replaced with an arbitrary short string (say, d45e3r) until you perform the analysis, and then replace these short strings with your original names.
If you're using Phylip programs with DNA or protein sequences, you can do a full phylogenetic workflow using the BIRCH system. BioLegato, the graphic user interface for BIRCH, automatically translates sequence names to a short random name compatible with Phylip, and then restores the names in the output. Name translation is done by uniqid.py. An example of Phylip output with long sequence names is shown below:
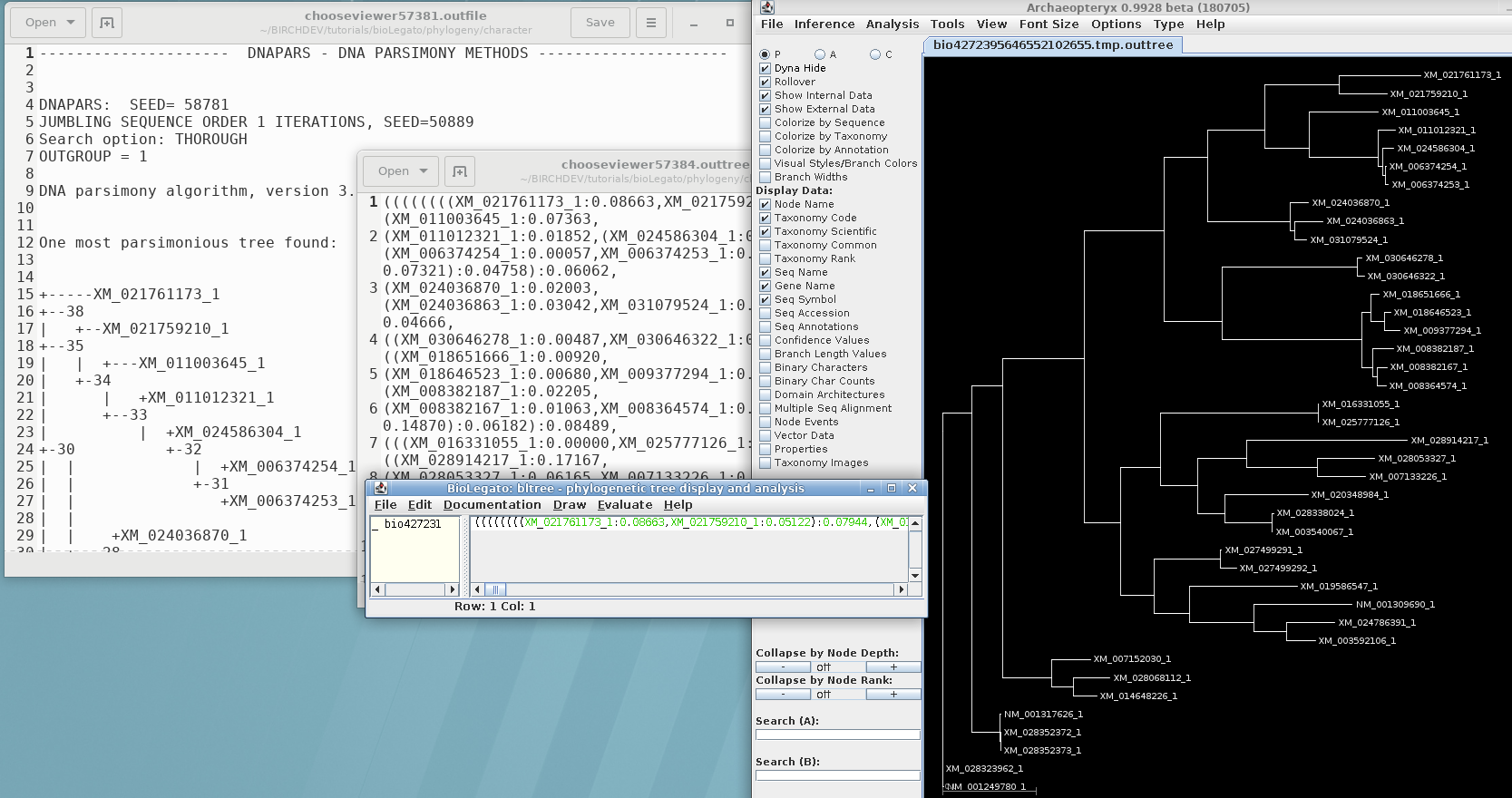
Further examples can be see no the BioLegato tutorials page under the heading "Phylogeny".
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Thanks will look into it.