Hi,
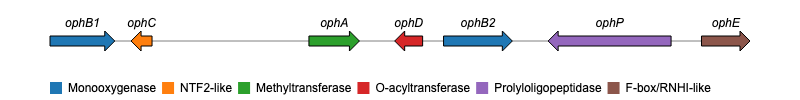
I am wondering with which tool I can plot the gene clusters like BGCs. Like figure below:
Or like Fig S1E:
https://ars.els-cdn.com/content/image/1-s2.0-S0092867414008265-figs1_lrg.jpg
Thanks
Hi,
I am wondering with which tool I can plot the gene clusters like BGCs. Like figure below:
Or like Fig S1E:
https://ars.els-cdn.com/content/image/1-s2.0-S0092867414008265-figs1_lrg.jpg
Thanks
There are really A LOT of tools for this task, commercial, free open source, Java, R, Python, C++ libraries. See here f.e.: What Tools/Libraries Do You Use To Visualize Genomic Feature Data? most of these are still existing and working. Depends on your input format your requirements, how many clusters to draw, and how much time you want to trade for your image to look exactly as you have in mind.
To not get lost in all the options, try UGENE first if you have a GenBank file and few clusters to plot. If it doesn't work for you, knowing the reasons why will most likely give you a better understanding of what you need.
Possibly
https://cran.r-project.org/web/packages/gggenes/vignettes/introduction-to-gggenes.html
http://genoplotr.r-forge.r-project.org/
https://github.com/thackl/gggenomes
I maintain a long list of tools here, there may be others :) https://cmdcolin.github.io/awesome-genome-visualization/?latest=true
I have made an R package called geneviewer that is designed for drawing gene arrow maps like the example below. Have a look at the package website and github
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.
How to visualize gene structure of a (prokaryotic) locus?