Can someone please explain me the logic behind identifying genes present within 50KB, 100KB and 500KB (both side) of a SNP locus ? How does the SNP affect the function of the genes present within the above mentioned windows?
Several studies plot the density (or effect size) of eQTLs by distance to gene TSS; such as:
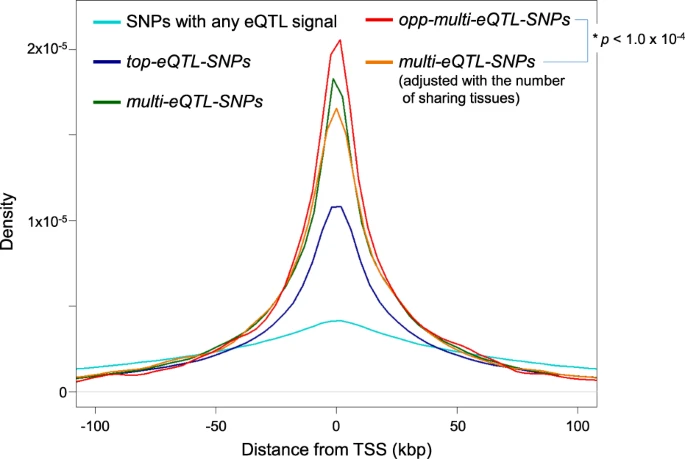
As you can see, these desnities are highly peaked around the TSS, but are heavy tailed. The reason to go further than 100kb or 250kb is to capture long-range eQTLs that can arise from chromatin looping -- i.e., bringing opposite sides of topologically associating domains (TADs) into close proximity. (Sensitivity)
A reason not to go further than 100kb or 250kb is power; by incorporating more genes (or more SNPs) into your analysis, with insufficient data you necessarily lose power to detect what you're looking for, and run the risk of false-negatives (and potentially false-positives as well) by diluting true signal. (Specificity)
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.




Thank you for your answer !